Abstract
The first complete chloroplast genome (cpDNA) sequence of Michelia chapensis was determined from Illumina HiSeq pair-end sequencing data in this study. The cpDNA is 160,138 bp in length, contains a large single-copy region (LSC) of 88,164 bp and a small single-copy region (SSC) of 18,824 bp, which were separated by a pair of inverted repeats (IR) regions of 26,575 bp. The genome contains 132 genes, including 87 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. Further phylogenomic analysis showed that M. chapensis is closed to M. maudiae and M. wilsonii in the tribe Michelieae.
Michelia chapensis Dandy is the species of the genus Michelia Linnaeus within the family Magnoliaceae, native in Fujian, Guangdong, Guangxi, Guizhou, Hongkong, Hunan, Jiangxi, Xizang, Yunnan and Zhejiang of China and Vietnam (Sima and Lu Citation2009; Shao et al. Citation2011). It is the key protected rare and endangered plant in China (Liu et al. Citation2004). Michelia chapensis Dandy is widely used in landscaping, and a precious fast-growing broad-leaved timber species (Lai Citation2008). The timber has the advantages of straight texture, small density, even structure (Li and Li Citation1999). It is an ideal alternative tree species for Pinus massoniana Lambert, Cunninghamia lanceolata (Lambert) Hooker and other coniferous tree species (Liu Citation2007). However, there has been no genomic study on M. chapensis Dandy.
Herein, we reported and characterized the complete M. chapensis Dandy plastid genome. The GenBank accession number is MN897730. One individual of M. chapensis Dandy was collected from Kunming Arboretum, Yunnan Academy of Forestry & Grassland Science, Yunnan Province of China (25°14′58″ N, 102°75′33″E). The sheets of vouchered specimen, Y. K. Sima 98270, are stored at the herbaria, YAF and YCP. DNA was extracted from its fresh leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA).
Paired-end reads were sequenced using Illumina HiSeq system (Illumina, San Diego, CA). In total, about 9.1 million high-quality clean reads were generated with adaptors trimmed. Aligning, assembly, and annotation were conducted by CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and GENEIOUS v 11.0.5 (Biomatters Ltd, Auckland, New Zealand). To confirm the phylogenetic position of M. chapensis Dandy, other 12 species of the tribe Michelieae from NCBI were aligned using MAFFT v.7 (Katoh and Standley Citation2013). The auto algorithm in the MAFFT alignment software was used to align the 15 complete genome sequences and the G-INS-i algorithm was used to align the partial complex sequences. The maximum likelihood (ML) bootstrap analysis was conducted using RAxML (Stamatakis Citation2006); bootstrap probability values were calculated from 1000 replicates. Based on the system of Magnoliaceae by Sima and Lu (Citation2012), two species of the tribe Magnolieae, Magnolia grandiflora Linnaeus (JN867587) and Lirianthe delavayi (Franchet) N. H. Xia & C. Y. Wu (MN780910) were served as the out-group.
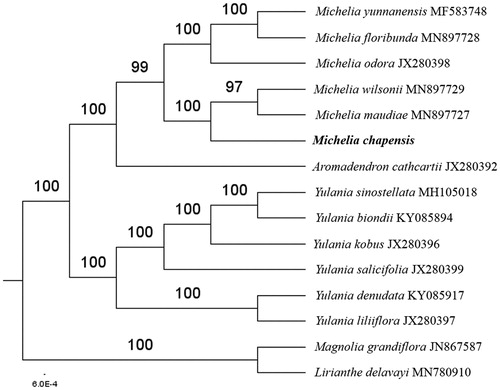
The complete M. chapensis Dandy plastid genome is a circular DNA molecule with the length of 160,138 bp, contains a large single-copy region (LSC) of 88,164 bp and a small single-copy region (SSC) of 18,824 bp, which were separated by a pair of inverted repeats (IR) regions of 26,575 bp. The overall GC content of the whole genome is 39.2%, and the corresponding values of the LSC, SSC, and IR regions are 37.9%, 34.2%, and 43.2%, respectively. The plastid genome contained 132 genes, including 87 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. Phylogenetic analysis showed that M. chapensis Dandy close to M. maudiae Dunn and M. wilsonii Finet & Gagnepain in the tribe Michelieae and the tribe Michelieae including the three genera, Aromadendron Blume, Yulania Spach and Michelia Linnaeus, is a nomophyletic group under the system of Magnoliaceae (Sima and Lu Citation2012) (). The determination of the complete plastid genome sequences provided new molecular data to illuminate the tribe Michelieae in Magnoliaceae evolution.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Lai D. 2008. Comparison of fiber morphology of Michelia tsoi wood between plantation and natural forest. Subtrop Agric Res. 4(2):89–90.
- Li YY, Li DX. 1999. Conservation value and exploitation foreground of the magnoliaceae plants in Yunnan. J Beijing Univ. 21(3):29–35.
- Liu HT. 2007. A study on the phenophase of Michelia chapensis. J Fujian Forestry Sci Tech. 34(2):112–114.
- Liu KW, Su Y, Hou BQ. 2004. Research on the characteristics and dynamics of Michelia chapensis communities. J Central South Univ. 21(4):47–50.
- Shao WH, Jiang JM, Luan QF, Liu ZX. 2011. A study on variation of growth traits in young Michelia chapensis forest and prove-nances selection. Acta Agricult Univ Jiangxi. 33(4):701–706.
- Sima YK, Lu SG. 2009. Flora of Southeast Yunnan. Vol. 1, Magnoliaceae. Kunming: Yunnan Science & Technology Press.
- Sima YK, Lu SG. 2012. A new system for the family Magnoliaceae. In: Xia NH, Zeng QW, Xu FX, Wu QG, editors. Proceedings of the second international symposium on the family Magnoliaceae. Wuhan: Huazhong University Science & Technology Press; p. 55–71.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.