Abstract
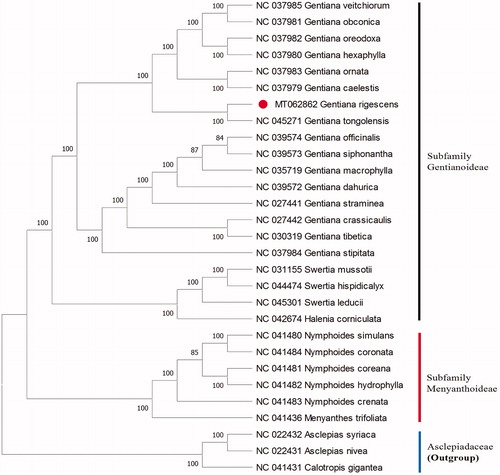
Gentiana rigescens Franch. ex Hemsl. (Gentianaceae) is an endangered medicinal plant endemic to the southwest of China. The chloroplast (cp) genome of G. rigescens was 146,891 bp in length, with a large single-copy (LSC) region of 79,377 bp, a small single-copy (SSC) region of 17,026 bp, and a pair of inverted repeats (IRs) of 25,244 bp, forming a typical quadripartite structure. A total of 130 genes were annotated from the cp genome of G. rigescens, including 85 protein-coding genes, 37 tRNA genes, and 8 ribosomal RNA genes. Phylogenetic analysis indicated that G. rigescens was closely related to G. tonggolensis with strong bootstrap values belonging to the subfamily Gentianoideae.
Gentiana rigescens is a perennial species of the family Gentianaceae, which is mainly distributed in the Yunnan, Sichuan, Guizhou, Hunan, and Guangxi provinces in China (Pan et al. Citation2015). The root of G. rigescens has been commonly used as a traditional Chinese medicine, for the treatment of convulsion, rheumatism, and jaundice, etc. (Qi et al. Citation2017). Due to the over-exploitation of wild resources and narrow distribution, the wild populations of G. rigescens are severely threatened (Zhu et al. Citation2019). To date, the previous studies were mainly focused on its resource, chemical composition, genetic diversity, and pharmacodynamics (Xu et al. Citation2009; Wu et al. Citation2017), rarely involved in its genomes. Here, we assembled and characterized the complete cp genome of G. rigescens and revealed its phylogenetic relationships for the first time, which will contribute to further studies on its genetic research and resource utilization.
Fresh leaves of G. rigescens were collected from Cangshan mountain (25°41′4″N, 100°08′31″E) of Dali counties, Yunnan province, China. The voucher specimen was deposited in the herbarium of Dali University (HBGP0420). Total DNA was isolated using the DNeasy plant mini kit (QIAGEN). Paired-end reads were sequenced by using Illumina NovaSeq platform (San Diego, CA). Approximately 8.5 Gb of raw data (56,765,498 reads) were assembled by NOVOPlasty v3.7 (Dierckxsens et al. Citation2017), and the assembled cp genome was annotated by GeSeq (Michael et al. Citation2017). Finally, the annotated cp genome was submitted to the GenBank (accession number MT062862).
The cp genome was 146,891 bp in length, with 37.83% overall GC content and exhibited a typical quadripartite structure, with a pair of IRs (inverted repeats) of 25,244 bp separated by a small single-copy (SSC) region of 17,026 bp and a large single-copy (LSC) region of 79,377 bp. It contained 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 ribosomal RNA genes. In order to reveal the phylogenetic position of G. rigescens, a phylogenetic analysis was performed based on 29 complete cp genomes of Gentianaceae and three taxa (NC 022431, NC 022432, and NC 041431) as out-group. The sequences were aligned by MAFFT v7.307 (Katoh and Standley et al. Citation2013), and the maximum likelihood (ML) bootstrap analysis was conducted using RAxML (Stamatakis Citation2014). Bootstrap probability values were calculated from 1000 replicates. As a result, phylogenetic analysis indicated that G. rigescens was closely related to G. tonggolensis with strong bootstrap values belonging to the subfamily Gentianoideae (). The cp genome resource may provide valuable guidelines for the management and utilization of G. rigescens.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7 improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Michael T, Pascal L, Tommaso P, Ulricht-Jones ES, Axel F, Ralph B, Stephan G. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. W1(45):W6–W11.
- Pan Y, Zhang J, Shen T, Zuo ZT, Jin H, Wang YZ, Li WY. 2015. Optimization of ultrasonic extraction by response surface methodology combined with ultrafast liquid chromatography-ultraviolet method for determination of four iridoids in Gentiana rigescens. J Food Drug Anal. 3(23):529–537.
- Qi L, Zhang J, Zuo Z, Zhao Y, Wang Y, Hang J. 2017. Determination of iridoids in Gentiana rigescens by infrared spectroscopy and multivariate analysis. Anal Lett. 2(50):389–401.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 9(30):1312–1313.
- Wu Z, Zhao Y, Zhang J, Wang Y. 2017. Quality assessment of Gentiana rigescens from different geographical origins using FT-IR spectroscopy combined with HPLC. Molecules. 7(22):1238.
- Xu M, Yang CR, Zhang YJ. 2009. Minor antifungal aromatic glycosides from the roots of Gentiana rigescens (Gentianaceae). Chinese Chem Lett. 10(20):1215–1217.
- Zhu B, Luo X, Gao Z, Feng Q, Hu L, Weng Q. 2019. The complete chloroplast genome sequence of Dendrobium thyrsiflorum (Orchidaceae) and its phylogenetic analysis. Mitochondr DNA B. 2(4):4085–4086.