Abstract
This study was conducted to sequence, assemble, annotate, and characterize the complete chloroplast (cp) genome sequence of Ulmus lanceaefolia. The result showed that the cp genome was 158,652 bp in length with 35.63% GC content, including a large single-copy (LSC) region of 87,119 bp, a small single-copy (SSC) region of 18,697 bp and an inverted repeat sequence (IRa/IRb) region of 26,418 bp, which was a typical tetrad structure. We had identified a total of 132 genes, including 87 protein-coding genes, eight rRNA genes, and 37 tRNA genes. The phylogenetic trees were constructed based on the cp genome sequences of U. lanceaefolia and 12 plants in the NCBI database, and the phylogenetic positions of U. lanceaefolia in the Ulmaceae were identified.
Chloroplast genome, as the unique and relatively independent genome from nuclear genome of plants, has the characteristics of relatively small molecular mass, low nucleotide replacement rateand slow structural variation, etc. Consequently, they are increasingly used to resolve the problems of angiosperm phylogeny (Moore et al. Citation2010; Gao et al. Citation2019). Ulmaceae is an important plant family with approximately 16 genera and 230 species, widely distributed in temperate and tropical regions throughout the world, mainly in the northern temperate zone (Airy Show and Williss Citation1973). Nowadays, chloroplast genome and phylogenetic studies have been carried out on some plants of Ulmaceae, such as Ulmus davidiana, Ulmus laciniata (Zuo et al. Citation2017), but there is no more relevant report on U. lanceaefolia.
Ulmus lanceaefolia, alternate name yunnan elm, evergreen yunnan elm etc., is a large evergreen tree, reaching a height of 40 m, native to montane forests (500–1500 m elevation) in Yunnan Province, China. The experimental material were planted in Hebei Academic of Forestry and Grassland, Shijiazhuang, China (114°28′12″E, 38°08′23″N), and its herbarium is stored in the herbarium of Hebei Academic of Forestry and Grassland, File number is HAFG22U376. DNA extraction, chloroplast genome sequencing, assembly and annotation were completed in Beijing Zhongxing Bomai Technology Co., Ltd. Now, the DNA of U. lanceaefolia is stored in an ultralow-temperature refrigerator of Hebei Engineering Research Center for Trees, Hebei, China.
The cp genome of U. lanceaefolia was sequenced using the Illumina NovaSep platform, then the clean reads were assembled and spliced to obtain a closed circle genome. The total length of the cp genome is 158,652 bp with 35.63% overall GC content. A pair of IRa/IRb (inverted repeats) of 26,418 bp were separated by SSC (18,697 bp) and LSC (87,119 bp). The GC content in LSC, SSC, IRA, and IRb regions were 33.14%, 36.84%, 42.31%, and 38.67%, respectively. Totally 132 genes were identified, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Of these, six protein-coding genes, seven tRNA genes, and four rRNA genes were duplicated in the inverted repeat regions.
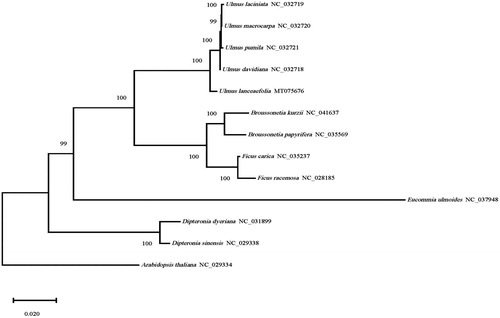
The phylogenetic tree was constructed based on the complete chloroplast sequences of U. lanceaefolia and 12 other plants species available in the NCBI database (including four Ulmus species,two Ficus species, two Broussonetia species, one Eucommia species, two Diteronia species, and a outgroup Arabidopsis) using maximum likelihood (ML) tree with 1000 bootstrap replicates. The study showed that (), five species in Ulmus were clustered into one branch, which were closely related to Morus plants. And the results of the branch clustering of the genus Ulmus were divided into ser. Lanceaefoliae (U. lanceaefolia), ser. Glabrae Moss (including U. laciniata, U. macrocarpa, and U. pumila), and ser. Nitentes Moss (U. davidiana). It is consistent with the conclusion about the classification of Ulmus in engler classification system. It was the first time to identify the phylogenetic position of U. lanceaefolia in Ulmaceae, and provided theoretical basis for molecular identification and resource exploitation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Airy Show HK, Williss JC. 1973. A dictionary of the flowering plant and ferns. 8th edn. Cambridge: Cambridge University Press.
- Gao L, Liu Y, Zhang D, et al. 2019. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun Biol. 2:278.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107(10):4623–4628.
- Zuo LH, Shang AQ, Zhang S, Yu XY, Ren YC, Yang MS, Wang JM. 2017. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: genome comparative and taxonomic position analysis. PLoS One. 12(2):e0171264–19.