Abstract
Citrus medica (C. medica), belonging to the family Rutaceae, possibly originated in north-east India and is now cultivated in subtropical and tropical regions. In this study, the assembled cp genome of C. medica exhibited a typical quadripartite cycle of 160,031 bp, composing of a pair of inverted repeats (IRs, 26,991 bp each) separated by a large single-copy (LSC, 87,476 bp) region and a small single copy (SSC, 18,573 bp) region. A total of 134 genes were annotated in this cp genome. Phylogenetic analysis showed that C. medica was closely related to Citrus limon (C. limon) and Citrus sinensis (C. sinensis).
Citrus medica, commonly known as citron, is one of the original citrus fruits from which all other citrus types developed through artificial hybridization or natural hybrid speciation (Wu et al. Citation2018). The genome of C. medica displays a high degree of genetic homozygosity because it is usually fertilized by self-pollination. Previous studies have demonstrated that C. medica is the male parent of any citrus hybrid rather than a female one, and the hybrids of citrons with other citrus are commercially prominent, notably lemons and many limes (Nicolosi et al. Citation2000; Carvalho et al. Citation2005; Barkley et al. Citation2006). Additionally, it has a series of applications in Asian cuisine, and also in traditional medicines, perfume, and for religious rituals and offerings. The complete chloroplast (cp) genome of C. medica was assembled and characterized herein to provide more evidence for the identification and application of citron.
The fresh leaves of C. medica were collected from Corsica, France (42°18′31.6′′N, 9°12′22.7′′E). The voucher specimen is now deposited at the Herbarium of Nanjing Forestry University (accession number: 20171017ivia_322). Genomic DNA was extracted from the fresh leaves and then sequenced using the Illumina Hiseq 2000 platform. The raw sequencing data were filtered and trimmed by fastp program (Chen et al. Citation2018), and then fed into NOVOPlasty version 3.7.2 (Dierckxsens et al. Citation2017) for assembly with the seed of psbA and reference genome sequence of Citrus sinensis (GenBank accession: NC_008334.1). The assembled genome was then annotated using PGA (Qu et al. Citation2019) and adjusted manually using Macvector v17.0.7. The complete cp genome was submitted to GenBank (accession number: MT106673).
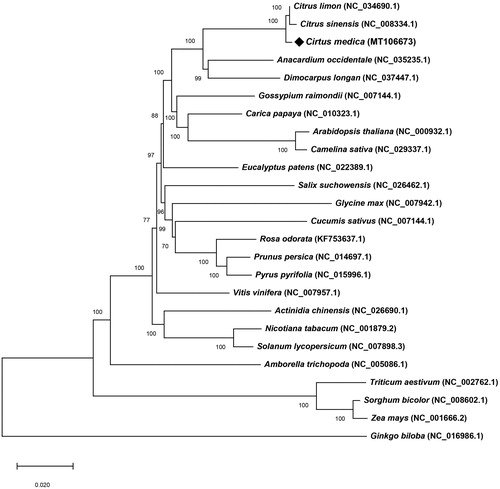
The cp genome of C. medica is 160,031 bp in length with the typical quadripartite structure of angiosperms, including 87,476 bp large single-copy (LSC) region and 18,573 bp small single-copy (SSC) region separated by a pair of 26,991 bp inverted repeat (IR) regions. The overall GC content of C. medica cp genome was 38.44%, which is higher than that of LSC (36.79%) and SSC (33.28%), but lower than IRs (42.91%). The C. medica cp genome contained 134 genes, including 89 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Twenty genes were found to contain one intron, including 12 protein-coding genes (rps16, atpF, rpoC1, rps12 × 2, petB, petD, rpl2 × 2, ndhB × 2, and ndhA), and 8 tRNA genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU × 2, and trnA-UGC × 2), and two genes contain two introns (ycf3 and clpP). Phylogenetic analysis was carried out using 73 conserved protein-coding genes with those of 25 plant cp genomes by neighbor-joining (NJ) method in MEGA X (Kumar et al. Citation2018). The phylogenetic tree showed that C. medica was clustered to other species in the Rutaceae family (Citrus limon and Citrus sinensis) and the family was evolutionarily close to Anacardium occidentale and Dimocarpus longan (). The study will provide more evidence for the identification and application of citron.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Barkley NA, Roose ML, Krueger RR, Federici CT. 2006. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet. 112(8):1519–1531.
- Carvalho R, Soares Filho WS, Brasileiro-Vidal AC, Guerra M. 2005. The relationships among lemons, limes and citron: a chromosomal comparison. Cytogenet Genome Res. 109(1–3):276–282.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Nicolosi E, Deng ZN, Gentile A, Malfa SL, Continella G, Tribulato E. 2000. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet. 100(8):1155–1166.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15(1):50.
- Wu GA, Terol J, Ibanez V, López-García A, Pérez-Román E, Borredá C, Domingo C, Tadeo FR, Carbonell-Caballero J, Alonso R, et al. 2018. Genomics of the origin and evolution of Citrus. Nature. 554(7692):311–316.