Abstract
The complete mitochondrial genome of bark beetle Scolytus schevyrewi is determined. This mitogenome is 15,891 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a control region (D-loop), with an A + T bias of 65.33%. Its gene order is similar to the mitogenomes of Coleoptera.
Scolytus schevyrewi (Coleoptera: Curculionidae) is an important pest of elms, its main host plants include many kinds of elms and it can spread the Dutch elm disease pathogens (Jacobi et al. Citation2013). In China, S. schevyrewi is distributed in Beijing, Hebei, Heilongjiang, Henan, InnerMongolia, Ninxia, Qinghai, Shaanxi, Shanxi and Xinjiang (Wood and Bright Citation1992; Bright and Skidmore Citation2002). In this study, the specimens of S. schevyrewi were collected from the elms in Yining, Xinjiang Province, China (N43°42′17″, E 81°10′15″; 9 August 2018). The voucher specimens are kept at the Laboratory of Invasion Biology, College of Agriculture, Jiangxi Agricultural University, Jiangxi, China. The specimen accession number is ZL20180809013.
The complete mitochondrial genome of S. schevyrewi (Genbank: MK636870)is 15,891 bp in length. The compositions of this mitochondrial genome include 28.15% A, 21.77% G, 12.90% C, and 37.18% T, and it has an A + T bias of 65.33%. The S. schevyrewi mitogenome contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and a control region (D-loop). The overall length of the 13 PCGs is 10,804 bp. Except for trnN lacking the TψC loop and trnS1 lacking the DHU arm and loop, the remaining tRNA genes all have typical clover-leaf structures. The length of the tRNA genes range from 55 to 69 bp. The 2 rRNA genes are 1053 bp and 764 bp, respectively.
Except for seven tRNA genes (trnF, trnY, trnC, trnQ, trnV, trnL1, trnP, trnH), and nad5, nad1, nad4L, nad4, rrnS, rrnL genes lie on the light strand (L-strand), the other genes are located on the heavy strand (H-strand). This gene order accords with the mitochondrial genomes arrangement of Coleoptera (Timmermans and Vogler Citation2012). All protein initiation codons are ATN. The standard termination codon TAG is used by nad3, atp8, nad1, cob and nad4, and the remaining genes use TAA as termination codon, except for cox2 has an incomplete termination codon with a single T.
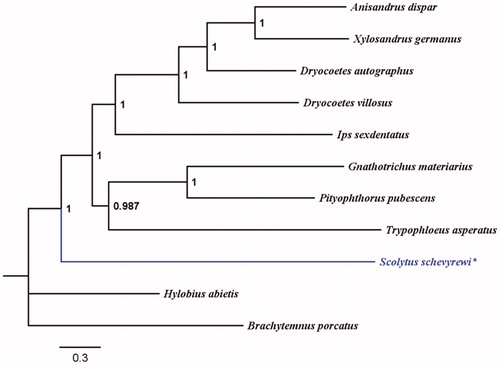
The MrBayes molecular phylogeny shown in is based on the 13 PCGs, 22tRNAs, and 2 rRNAs of S. schevyrewi and 8 other Scolytinae beetles is constructed using PhyloSuite (Ronquist et al. Citation2012; Katoh and Standley Citation2013; Lanfear et al., Citation2016; Zhang et al. Citation2019), Hylobius abietis and Brachytemnus porcatusare used as outgroups. This phylogenetic tree is similar to the phylogenetic tree of IQ-tree analysis (Nguyen et al. Citation2015). Scolytus schevyrewi and the branch including the other Scolytinae beetles cluster into a bigger branch. This result is similar to the previous phylogeny studies of Curculionoidea, the tribe Scolytini is placed outside a polytomy including the remaining species of Scolytinae (Gillett et al. Citation2014; Pistone et al. Citation2016). The complete mitochondrial genome of S. schevyrewi will provide useful information for the further studies about taxonomic classification, phylogenetic reconstruction, and planning control measures.
Figure 1. MrBayes phylogenetic tree based on the 13 PCGs, 22tRNAs and 2 rRNAs of Scolytus schevyrewi and 8 other Scolytinae beetles. Hylobius abietis and Brachytemnus porcatus are used as outgroups. GenBank accession numbers for each species: Anisandrus dispar NC036293, Xylosandrus germanus KX035202, Dryocoetes autographus NC036287, Dryocoetes villosus NC036282, Ips sexdentatus KX035215, Gnathotrichus materiarius NC036294, Pityophthorus pubescens NC036288, Trypophloeus asperatus NC036285, Scolytus schevyrewi MK636870, Hylobius abietis JN163954, Brachytemnus porcatus JN163960.
Acknowlegements
We thank Chengpeng Long (Zhejiang A&F University), Peng Liu (Anhui Agricultural University) for dataprocessing and guide.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Bright DE, Skidmore RE. 2002. A catalog of Scolytidae and Platypodidae (Coleoptera), supplement 2 (1995–1999). Ottawa: NRC Research Press.
- Gillett C, Crampton-Platt A, Timmermans M, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of Weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31(8):2223–2237.
- Jacobi WR, Koski RD, Negron JF. 2013. Dutch elm disease pathogen transmission by the banded elm bark beetle Scolytus schevyrewi. Forest Pathol. 43(3):232–237.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lanfear R. Frandsen PB. Wright AM. Senfeld T. Calcott B. 2016. PartitionFinder2: new methods for selecting partitioned models of evolution formolecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pistone D, Mugu S, Jordal BH. 2016. Genomic mining of phylogenetically informative nuclear markers in bark and Ambrosia beetles. PLOS One. 11(9):e0163529.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Timmermans M, Vogler AP. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol Phylogenet E. 63:229–304.
- Wood SL, Bright DE. 1992. A catalog of Scolytidae and Platypodidae (Coleoptera), Part 2: taxonomic index volume A and B. Great Basin Nat Mem. 13:1–1553.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2019. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
