Abstract
In this study, we sequenced and annotated the complete chloroplast genome of Codium fragile (GenBank accession number: MN853876) .The total length of the mitogenome is 83,788 bp including 78 protein encoding genes, 26 tRNA genes and 3 rRNA. The C+G composition of the complete chloroplast genome of Codium fragile is 27.63% and lower than A+T. The phylogenetic tree which based on Core gene shows Codium fragile clustered into Condium clade and had close genetic relationship with algae Condium arabicum. The data will increase the basic information of macroalgae phylogenetic research and can help to better understand the phylogenetic status of Codium fragile.
Codium fragile belongs to Eukaryon, Plantae, Chlorophyta, Chlorophyceae, Codium orders, Codiaceae, Codium, which is distributed around the world (Watanabe et al. Citation2009). It has a large number of special polysaccharide structure and the effects of anti-tumor (Wijesekara et al. Citation2011), anti-virus and anti-coagulation (Ciancia et al. Citation2007). As an important algae resources, Codium fragile should be studied in different aspects. The chloroplast genome contains more protein-coding genes, which plays an important role in plant phylogeny and genetic diversity (Cai et al. Citation2017, Citation2018; Cremen et al. Citation2018). Large number of chloroplast genomes of green algae have been published according to NCBI (Cai et al. Citation2017). However, the chloroplast genome of Codium fragile has never been reported before. So in this study, we determined the complete chloroplast genome sequence of Codium fragile.
Codium fragile was collected from Qinhunangdao coastal area (39°49′54.69″N, 119°31′30.50″E) cultured in laboratory with VSE medium at 20°Cunder a light intensity of 100 μmol−2 s−1 (Zhou et al. Citation2016a, Citation2016b). The specimens were preserved at the Marine Ecology Research Center of the First Institute Oceanography, Ministry of Natural Resources in Qingdao (Accession number: CSZ06). The shape of genome of Codium fragile is a double-chain closed loop with GenBank accession number MN853876. The content of C + G is 74.34%. The complete chloroplast genome sequence is 83,788 bps long. There are 78 protein-coding genes in the genome, including 15 psb genes, 11 rps genes, 10 rpl genes, 6 psa, atp and ycf genes, 5 pet and rpo genes, 4 chl genes, 2 cys genes. Besides, there are some genes which are contained only once in the genome, such as acc, ccs, cem, clp, fts, inf, rbc and tuf genes. For non-coding genes, 26 tRNAs, one rrl, rrs and rrn5 are included. All the coding genes begin with ATG except for accD, atpF, chlB, psbK, rpoB and rps9 with ATT. As to termination codons, cemA, chlB, psbZ and ycf12 terminate with TAG, while accD, atpI, rpl32, rps2 and ycf47 have the termination TGA. The rest 60 genes end with TAA.
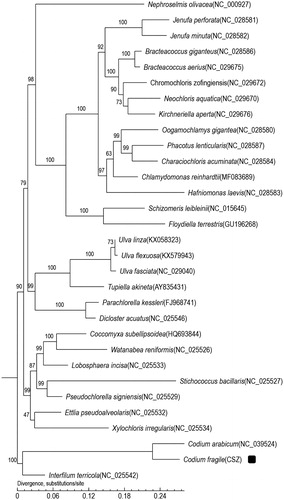
This is the first record of chloroplast genome of Codium fragile. The phylogenetic relationship of Codium fragile with other species were estimated using the phylogenetic tree constructed based on core gene of Codium fragile and other 30 species. The phylogenetic tree () showed Codium fragile clustered into Codium clade and had close genetic relationship with algae Condium arabicum.
Disclosure statement
We declare that we have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cai CE, Wang LK, Jiang T, Zhou LJ, He PM, Jiao BH. 2018. The complete mitochondrial genomes of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta). Conserv Genet Resour. 10(3):415–418.
- Cai CE, Wang LK, Zhou LJ, He PM, Jiao BH. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One. 12(9):e0184196.
- Ciancia M, Quintana I, Vizcargüénaga MI, Kasulin L, Dios A, Estevez JM, Cerezo AS. 2007. Polysaccharides from the green seaweeds Codium fragile and C.vermilara with controversial effects on hemostasis. Int J Biol Macromol. 41(5):641–649.
- Cremen MCM, Leliaert F, Marcelino VR, Verbruggen H. 2018. Large diversity of non-standard genes and dynamic evolution of chloroplast genomes in siphonous green algae (Bryopsidales, Chlorophyta). Genome Biol Evol. 10(4):1048–1061.
- Watanabe S, Metaxas A, Scheibling RE. 2009. Dispersal potential of the invasive green alga Codium fragile ssp. fragile. J Exp Mar Biol Ecol. 381(2):114–125.
- Wijesekara I, Pangestuti R, Kim SK. 2011. Biological activities and potential health benefits of sulfated polysaccharides derived from marine algae. Carbohydr Polym. 84(1):14–21.
- Zhou LJ, Wang LK, Zhang JH, Cai CE, He PM. 2016a. Complete mitochondrial genome of Ulva linza, one of the causal species of green macroalgal blooms in Yellow Sea, China. Mitochondr DNA B. 1(1):31–33.
- Zhou LJ, Wang KL, Zhang JH, Cai CE, He PM. 2016b. Complete mitochondrial genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondr DNA B. 1(1):76–78.