Abstract
Rubus Linnaeus has long been deemed as one of the most diverse genera in family Rosaceae as well as an extremely challenging group with complicated intra-and-inter relationships at species level. These species are characterized by abundant diversity due to the tendency for hybridization, polyploidization, and agamospermy. There is a great deal of disagreement as to the identification, division of sections/subsections and phylogenetic history among a part of researchers. Hence, we assembled the complete chloroplast genome of Rubus cochinchinensis (MN913339) with 156,235 bp in total, composed of one large single copy-region (LSC, 85,845 bp), a small single-copy region (SSC, 18,848 bp) and a pair of inverted repeats (IRs, 51,542 bp). This genome contained 129 genes, including 8 rRNAs, 36 tRNAs, and 85 protein-coding genes and corporate CG content was 37.18%. Phylogenetic analysis provided a reliable basis for the divisions of tribes in the Rosaceae.
Rubus Linnaeus diffusely known as Raspberry, is a diverse population belonging to the subfamily Rosoideae with global distribution in all climates except Antarctica (Focke Citation1910a, Citation1911b, Citation1911c). The various ploidy level of this complex population ranges from diploid to dodecaploid, and all the benefits come from hybridization, polyploidization, and agamospermy. A portion of species, such as Rubus alceifolius, having large variations in main identification indicators, push the challenging limits of plant taxonomists (Alice and Campbell Citation1999). It contains approximately 740 species in the world (Gu et al. Citation1993). There are 201 species, 98 varieties distributing in China (Lu Citation1983) with abundant germplasm resources in the wild. These will provide a significant parent for breeders to select a hybrid combination with high heterosis. Thus, we explained the cytoplasm genome of Rubus cochinchinensis to apply a utility molecular data set and an elementary reference for future study.
Young R. cochinchinensis leaf samples were collected from natural populations in Hannan province, China. The voucher specimen (ID: RCOC-17602-HN02) was trapped in the Sun Yat-sen University Herbarium (SYS). The molecular sample was submitted to Jierui Bio-Technique Co. Ltd. for whole-genome pair-end sequencing base on the Illumina platform.
For the first step, the company’s feedback ‘clean_data’ sets (5.7 Gb) were aligned to the reference genome (Rubus leucanthus (MK105853.1)), which was assembled before by our team members and an intact circle genome was obtained via GetOrganelle v1.6.2 (Jin et al. Citation2018). In addition, Plann (Huang and Cronk 2008) was primarily used for the bulk of gene annotations and the rRNAs were marked by Geseq – Annotation of Organellar Genomes (https://chlorobox.mpimp-golm.mpg.de/geseq.html), an online annotation toolkit.
The complete chloroplast genome of Rubus cochinchinenisis (MN913339) was 156,235 bp to total length and consists of a large single-copy region (LSC) of 85,845 bp, a small single-copy region (SSC) of 18,848 bp and two inverted repeats (IRA and IRB) of 25,771 bp each. The chloroplast genome circle were joined end to end in the order of ‘LSC-IRA-SSC-IRB.’ The total CG content of the cytoplasm genome was 37.18% and LSC, SSC, and IRs, respectively, accounted for 54.95%, 12.06%, and 33% of the whole genome. It contained 8 rRNAs, 36 tRNAs, and 85 protein-coding genes. It is worth reminding that, the missing gene partition occurred at both LSC and IRs regions. The gene trnH-GUG and ycf1 dropped a single section for each gene, instead of the conventional paired structure of the reference genome.
To confirm the phylogeny position of R. cochinchinensis in Rosaceae, 21 representative species of diverse tribes within Rosaceae were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/). Siraitia grosvenorii (MK818498.1) was specified as one outgroup taxon. Using MAFFT v.7.271 (Katoh and Standley Citation2013), a total of 22 sequences were aligned and jmodeltest v.2.1.10 (David Citation2008) was applied for the model test. Eventually, the phylogenetic maximum-likelihood (ML) tree () was structured by means of RAxML v.8.2.12 (Stamatakis Citation2014).
Figure 1. Maximum-likelihood tree of Rosaceae base on complete chloroplast genomes, Bootstrap support values are shown next to the nodes. Tribes A ∼ E respectively represented as following: Potentilleae, Roseae, Rubeae, Maleae and Amygdaleae.
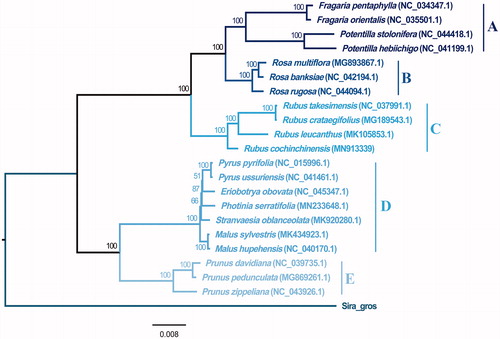
The phylogeny analysis indicated that R. cochinchinensis was placed in the Rubeae tribe, announcing prominent diversity compared with the other five tribes. The monophyletic group of these five tribes was well supported by ML tree generated from chloroplast genome data, which reported a common ancestor among these five tribes on account of molecular evidence.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Alice LA, Campbell CS. 1999. Phylogeny of Rubus (Rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am J Bot. 86(1):81–97.
- David P, 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 7:7.
- Focke WO. 1910a. Species Ruborum. Monographiae generis rubi prodromus part I. New York, NY: Stuttgart, E. Schweizerbart.
- Focke WO. 1911b. Species Ruborm. Monographiae generis rubi prodromus part II. New York, NY: Stuttgart, E. Schweizerbart. p. 121–223.
- Focke WO. 1911c. Species Ruborm. Monographiae generis rubi prodromus part II. New York, NY: Stuttgart, E. Schweizerbart. p. 224–498.
- Gu Y, Zhao CM, Jin W, Li WL, 1993. Evaluation of Rubus germplasm resources in China. Acta Hortic. 352:317–324.
- Huang DI, Cronk QCB, 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lu LD, 1983. A study on the genus Rubus of China. J Syst E. 21:13–25.
- Stamatakis A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
