Abstract
Rhopalosiphum nymphaeae (Linnaeus, 1761) is a heteroecious polyphagous aphid, which can survive underwater. We have determined mitochondrial genome of R. nymphaeae collected in Korea. The circular mitogenome of R. nymphaeae is 15,594 bp including 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 874 bp. The base composition was AT-biased (84.3%). Phylogenetic trees present that genus Rhopalosiphum is polyphyletic, requiring more investigation of mitochondrial genomes.
Water lily aphid, Rhopalosiphum nymphaeae (Linnaeus, Citation1761) is a heteroecious polyphagous aphid having Prunus species as winter hosts (Blackman and Eastop Citation2008) and inhabiting numerous wetland and water-dwelling plant species in summer (Atousa et al. Citation2015). This aphid is able to survive underwater (Holman Citation2009; Blackman and Eastop Citation2015). Its host list includes species of 45 plant families (Holman Citation2009). It has been reported as a pest (Hance et al. Citation1994) but has also been used to control water weeds in California (Oraze and Grigarick Citation1992). Till now there is no available complete mitochondrial genome of Rhopalosiphum genus.
Here, we presented mitochondrial genome collected in Korea (36°47'46.3"N 126°08'55.0"E; voucher was deposited in in Gyeongsang National University, Korea accession number: Coll#WH101). DNA was extracted using DNeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina HiSeq2000 (Macrogen Inc., South Korea) were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17, and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitochondrial genome based on that of Aphis gossypii (MN102349; Park, Jung, et al. Citation2019).
R. nymphaea mitochondrial genome (GenBank accession is MN943499) is 15,594 bp. Its nucleotide composition is AT-biased (A + T is 84.3%) and contains 13 protein-coding genes, two rRNAs, and 22 tRNAs. The control region, presumably corresponding to single largest non-coding AT-rich region (874 bp, A + T is 87.6%), is shorter than those of A. gossypii (Park, Jung, et al. Citation2019), Paracolopha morrisoni (Lee et al. Citation2019), and Myzus persicae. Gene order of R. nymphaea mitogenomes is identical to those of Aphididae species.
LR722139 sequence originated from Nymphaea colorata genome (Zhang et al. Citation2020) presented high nucleotide similarity with our mitogenome with two times duplicated regions. After removing repeat regions, 5 SNPs and 339 INDELs are identified. Most of INDELs are from 233-bp insertion in control region. Because of INDELs on genic region, this sequence should be corrected in near future. Number of SNPs and INDELs is smaller than those of Chilo suppressalis (Kwon, Kim, et al. Citation2019) and Spodoptera frugiperda (Seo, Lee, et al. Citation2019) and larger than those of Nilaparvata lugens (Choi et al. Citation2019; Kwon, Min, et al. Citation2019), Laodelphax striatellus (Park, Jung, et al. Citation2019; Seo, Jung, et al. Citation2019), Aphis gossypii (Park, Xi, et al., Citation2019), Stegobium paniceum (Park et al., under review), and Hipparchia autonoe (Lee et al., in preparation).
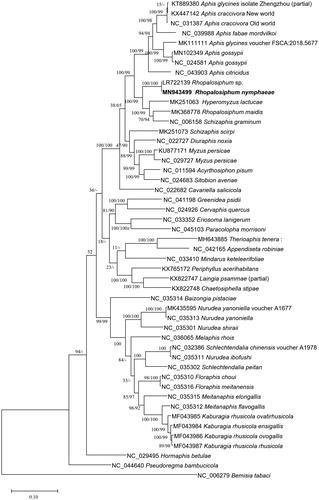
We inferred the phylogenetic relationship with 49 Aphididae mitogenomes including one outgroup species, Bemisia tabaci (Tay et al. Citation2016). Neighbor joining (10,000 bootstrap repeats) and maximum likelihood (1,000 bootstrap repeats) phylogenetic trees were constructed using MEGA X (Kumar et al. Citation2018) based on multiple sequence alignment of adjusted complete mitogenomes by MAFFT 7.450 (Katoh and Standley Citation2013). Phylogenetic trees present that Rhopalosiphum genus is polyphyletic with high bootstrap values and LR722139 is similar to our mitogenome ().
Figure 1. Neighbor joining (10,000 bootstrap repeats) and maximum likelihood (1,000 bootstrap repeats) phylogenetic trees of 48 insect species in the family Aphididae, R. nymphaea (MN943499 and trimmed LR722139 sequence), Rhopalosiphum maidis (MK368778), two Aphis gossypii (MN102349 and NC_024581), Myzus persicae (NC_029727 and KU877171), Acyrthosiphon pisum (NC_011594), Sitobion avenae (NC_024683), Diuraphis noxia (NC_022727), Cavariella salicicola (NC_022682), Schizaphis graminum (NC_006158), Aphis fabae mordvilkoi (NC_039988), Aphis craccivora (NC_031387 and KX447142), Aphis citricidus (NC_043903), Aphis glycines (KT889380 and MK111111), Chaetosiphella stipae (KX822748), Therioaphis tenera (MH643885), Pseudoregma bambucicola (NC_044640), Schizaphis scirpi (MK251073), Hyperomyzus lactucae (MK251063), Hormaphis betula (NC_029495), Cervaphis quercus (NC_024926), Greenidea psidii (NC_041198), Eriosoma lanigerum (NC_033352), Paracolopha morrisoni (NC_045103), Periphyllus acerihabitans (KX765172), Laingia psammae (KX822747), Baizongia pistaciae (NC_035314), Nurudea yanoniella (NC_035313 and MK435595), Nurudea shiraii (NC_035301), Nurudea ibofushi (NC_035311), Schlechtendalia chinensis (NC_032386), Schlechtendalia peitan (NC_035302), Melaphis rhois (NC_036065), Schlechtendalia elongallis (NC_035315), Nurudea choui (NC_035310), Nurudea meitanensis (NC_035316), Schlechtendalia flavogallis (NC_035312), Kaburagia rhusicola ovatirhusicola (MF043985), Kaburagia rhusicola ensigallis (MF043984), Kaburagia rhusicola ovagallis (MF043986), Kaburagia rhusicola rhusicola (MF043987), Mindarus keteleerifoliae (NC_033410), Appendiseta robiniae (NC_042165), and Bemisia tabaci (NC_006279) as an outgroup. Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Atousa F-H, Jalaeian M, Mehrparvar M. 2015. First report of Rhopalosiphum nymphaeae (L.) (Hem.: Aphididae) on Azolla filiculoides from Iran and its male formation on secondary host plant. J Crop Prot. 4(4):557–561.
- Blackman RL, Eastop VF. 2008. Aphids on the world’s herbaceous plants and shrubs, 2 volume set. Chichester: John Wiley & Sons.
- Blackman RL, Eastop VF. 2015. Aphids on the World’s Plants: an online identification and information guide; [accessed 2015 April 19]. http://www.aphidsonworldsplants.info/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Choi NJ, Lee B-C, Park J, Park J. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in China (Hemiptera: Delphacidae): investigation of intraspecies variations between countries. Mitochondrial DNA Part B. 4(1):1677–1678.
- Hance T, Nibelle D, Lebrun P, Impe G, Hove C. 1994. Selection of Azolla forms resistant to the water lily aphid, Rhopalosiphum nymphaeae Life history of Rhopalosiphum nymphaeae. Entomologia Experimentalis et Applicata. 70(1):11–17.
- Holman J. 2009. Host plant catalog of aphids. Palaearctic region. The Netherlands: Springer.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete chloroplast genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: low genetic diversity between Korea and Japan. Mitochondrial DNA Part B. 4(1):959–960.
- Kwon W, Min J, Kim Y, Park J. 2019. The complete chloroplast genome of Reboulia hemisphaerica (L.) Raddi (Aytoniaceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1459–1460.
- Lee J, Park J, Lee H, Park J, Lee W. 2019. The complete mitochondrial genome of Paracolopha morrisoni (Baker, 1919) (Hemiptera: Aphididae). Mitochondrial DNA Part B. 4(2):3037–3039.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Linnaeus, Carolus. 1761. Fauna Suecica sistens animalia Sueciae regni: Mammalia, Aves, Amphibia, Pisces, Insecta, Vermes. Distributa per classes et ordines, genera et species, cum differentiis specierum, synonymis auctorum, nominibus incolarum, locis natalium, descriptionibus insectorum. Editio altera. 578 PP.
- Oraze MJ, Grigarick AA. 1992. Biological control of ducksalad (Heteranthera limosa) by the waterlily aphid (Rhopalosiphum nymphaeae) in rice (Oryza sativa). Weed Sci. 40(2):333–336.
- Park J, Jung JK, Ho Koh Y, Park J, Seo BY. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Xi H, Kim Y, Park J, Lee W. 2019. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula. Mitochondrial DNA Part B. 4(2):3007–3009.
- Seo BY, Jung JK, Ho Koh Y, Park J. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a southern part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2242–2243.
- Seo BY, Lee G-S, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA Part B. 4(2):3918–3920.
- Tay W, Elfekih S, Court L, Gordon K, De Barro P. 2016. Complete mitochondrial DNA genome of Bemisia tabaci cryptic pest species complex Asia I (Hemiptera: Aleyrodidae). Mitochondrial DNA Part a. 27(2):972–973.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang L, Chen F, Zhang X, Li Z, Zhao Y, Lohaus R, Chang X, Dong W, Ho SYW, Liu X, et al. 2020. The water lily genome and the early evolution of flowering plants. Nature. 577(7788):79–76.
- Zhao Q, Wang Y, Kong Y-M, Luo D. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC bioinformatics, BioMed Central. 12(Suppl 14):S2.
