Abstract
Two mitogenomes of Ulva lactuca Linnaeus, one from Florida, USA and another nearly complete mtDNA from Chile, had previously been sequenced. Here, the complete mitogenome of U. lactuca from Shantou, China was sequenced and compared with them. The circular-mapping mitogenome of U. lactuca was 62,021 bp in size and 407 bases longer than that from the USA. The average sequence identity in gene sequence regions was 99.8% among these three U. lactuca mtDNAs. Differences in genome size were mainly caused by duplication mutations and insertion/deletion mutations of short DNA sequences. Two intergenic regions (nad6-trnS and trnF-trnG) contained long tandem repeats which displayed a high level of variation in repeat sequences and copy numbers, indicating fast mutation rates in these regions. The phylogenomic analysis revealed that these three samples of U. lactuca formed a strongly supported clade. The mitogenome can be used to understand the phylogeography and population genetics in Ulva on a global scale.
Ulva is the most speciose genus in the order Ulvales (Ulvophyceae, Chlorophyta), and harbors approximately 130 species (Guiry and Guiry Citation2020). Ulva species are macroscopic chlorophytes that are widespread in marine and estuarine environments and display cryptic diversity and great morphological plasticity (Hayden and Waaland Citation2002). They can cause large-scale harmful green tides (Melton et al. Citation2015; Wang et al. Citation2019). Ulva lactuca Linnaeus, a littoral and sublittoral sea lettuce, has been widely consumed as a marine vegetable in East Asia (Tang et al. Citation2016). Currently, Ulva fasciata Delile is regarded as a synonym of U. lactuca Linnaeus (Hughey et al. Citation2019). Here, the algal thalli of U. lactuca were collected from Nan’ao Island (23°28′N, 117°06′E), Shantou, Guangdong, China, and its specimen was deposited in the collection of marine algae in KLMEES of IOCAS (Specimen number: 2016-ST-Ula-1). The complete mitochondrial genome of U. lactuca was sequenced and compared with two U. lactuca mtDNAs from Florida, USA (KT364296) and Chile (MH763013).
The circular-mapping mitogenome of U. lactuca (GenBank accession number: KU182748) was constructed and annotated using the same protocol as previously described (Liu et al. Citation2017). It was 62,021 bp in size, which was 407 bases longer than that from the USA (Melton and Lopez-Bautista Citation2016). The overall sequence identity was 98.4% between two complete mitogenomes from China and the USA. These three mtDNAs contained the same set of 68 genes including 29 protein-coding genes, 30 tRNA genes, 2 rRNA genes, four intronic orfs, and three conserved free-standing orfs, and four introns. The cox1 gene harbored three group I introns (1401, 1249, and 1109 bp), each of which encoded a putative LAGLIDADG homing endonuclease. The nad3 gene encompassed one group II intron (2481 bp), which encoded a putative reverse-transcriptase/maturase. The genome organization and gene order were identical in these three mtDNAs.
The average sequence identity in gene sequence regions was 99.8% among these three mtDNAs. Non-coding intergenic regions accounted for 32.88% of the genome from China, which was slightly higher than that from the USA (32.40%). Differences in genome size among three mtDNAs were mainly caused by duplication mutations and insertion/deletion mutations of short DNA sequences. The vast majority of these mutations occurred in non-coding regions. Two intergenic regions (nad6-trnS and trnF-trnG) contained long tandem repeats which displayed a high level of variation in repeat sequences and copy numbers, indicating fast mutation rates in these regions. Some intraspecific differences have been observed in the mitgenomes of Ulva pertusa and Ulva prolifera, due to intron gain/loss, insertion of foreign DNA fragments, and rearrangement events (Liu and Pang Citation2016; Zhou et al. Citation2016; Liu et al. Citation2017).
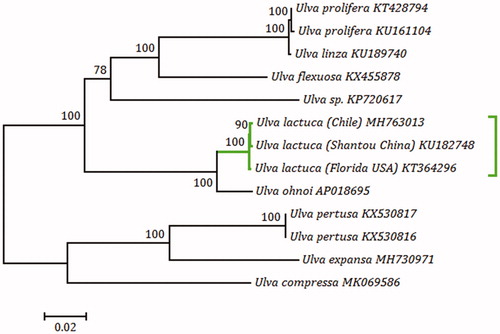
Based on nucleotide sequences of the 13 Ulva mitogenomes, a maximum likelihood (ML) tree was constructed using MEGA 7.0 (Kumar et al. Citation2016). The phylogenomic analysis revealed that three samples of U. lactuca formed a highly supported clade (100% bootstrap). The sample from China clustered more closely with that from Chile than with the sample from the USA (). The mitogenomic data will be a useful resource for our understanding of the phylogeography and population genetics in Ulva on a global scale.
Figure 1. Phylogenomic tree based on maximum-likelihood (ML) analysis of nucleotide sequences of the 13 known Ulva mitogenomes was conducted with 1000 bootstrap replicates using MEGA 7.0. Branch lengths were proportional to the amount of sequence change, which was indicated by the scale bar below the tree.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Guiry MD, Guiry GM. 2020. AlgaeBase. Galway, Ireland: National University of Ireland.
- Hayden HS, Waaland JR. 2002. Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. J Phycol. 38(6):1200–1212.
- Hughey JR, Maggs CA, Mineur F, Jarvis C, Miller KA, Shabaka SH, Gabrielson PW. 2019. Genetic analysis of the Linnaean Ulva lactuca (Ulvales, Chlorophyta) holotype and related type specimens reveals name misapplications, unexpected origins, and new synonymies. J Phycol. 55(3):503–508.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu F, Melton JT, Bi YP. 2017. Mitochondrial genomes of the green macroalga Ulva pertusa (Ulvophyceae, Chlorophyta): novel insights into the evolution of mitogenomes in the Ulvophyceae. J Phycol. 53(5):1010–1019.
- Liu F, Pang SJ. 2016. The mitochondrial genome of the bloom-forming green alga Ulva prolifera. Mitochondrial DNA Part A. 27(6):4530–4531.
- Melton JT, Leliaert F, Tronholm A, Lopez-Bautista JM. 2015. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLoS ONE. 10(4):e0121020.
- Melton JT, Lopez-Bautista JM. 2016. De novo assembly of the mitochondrial genome of Ulva fasciata Delile (Ulvophyceae, Chlorophyta), a distromatic blade-forming green macroalga. Mitochondrial DNA Part A. 27(5):3817–3819.
- Tang YQ, Kaiser M, Ruqyia S, Muhammad FA. 2016. Ulva lactuca and its polysaccharides: food and biomedical aspects. J Biol, Agric Healthcare. 6(1):140–151.
- Wang Y, Liu F, Liu X, Shi S, Bi Y, Moejes FW. 2019. Comparative transcriptome analysis of four co-occurring Ulva species for understanding the dominance of Ulva prolifera in the Yellow Sea green tides. J Appl Phycol. 31(5):3303–3316.
- Zhou LJ, Wang LK, Zhang JH, Cai C, He PM. 2016. Complete mitochondrial genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondrial DNA Part B. 1(1):76–78.
