Abstract
Caesalpinia sappan L. is a widely cultivated medicinal plant. As a first report in this genus, we presented the complete chloroplast genome of C. sappan using genome skimming of paired-end NGS data. The total length of the chloroplast genome was 160,192 bp and contained a typical quadripartite structure including one large single-copy (LSC) region (89,726 bp), one small single copy (SSC) region (18,358 bp) and a pair of inverted repeats (IR) regions (26,054 bp). The GC content of this genome was 36.0%. The whole genome contained 127 genes including 83 protein-coding genes, 36 tRNA genes, and eight rRNA genes. ML analysis revealed C. sappan was closely related to Haematoxylum brasiletto with strong bootstrap values belonging to the subfamily Caesalpinioideae of Leguminosae.
Caesalpinia sappan L. is a species in the subfamily Caesalpinioideae of Leguminosae, which is widely distributed in China, India, Laos, Malaysia, Myanmar, Africa, and America (Flora of China Editorial Committee of Chinese Academy of Sciences Citation2010). As an ideal landscape tree species, it has very high ornamental value, C. sappan is also a traditional medicinal plant, and its lignum is mainly used for anti-inflammatory and hypoglycemic treatment (Nirmal et al. Citation2015; Mueller et al. Citation2016). So far, most of the studies on Caesalpinia and its related genus have focused on chemical compositions, morphological taxonomy, and molecular phylogeny (Cai et al. Citation2012; Gagnon et al. Citation2013). However, there is no genomic report on C. sappan to date. Therefore, we sequenced the cp genomes of C. sappaan and characterized the genome features, which will provide useful information for further study of the genus (Park et al. Citation2019).
Fresh leaves of C. sappan were collected from the medicinal plant arboretum, Yunnan Branch, Institute of Medicinal Plant, Chinese Academy of Medical Science, Yunnan province of China (22°0′28″N, 100°47′17″E). The voucher specimen was deposited in the herbarium of Dali University (HBGP0153). The genomic DNA was extracted using the Plant Genomic DNA kit (Tiangen, Beijing, China), and sequenced using the Illumina NovaSeq system (Illumina, San Diego, CA). Denovo genome assembly was conducted by GetOrganelle (Jin et al. Citation2018) and annotated using GeSeq with default sets (Michael et al. Citation2017). The complete chloroplast genome of C. sappan was submitted to the GenBank database (Accession Number: MN933929).
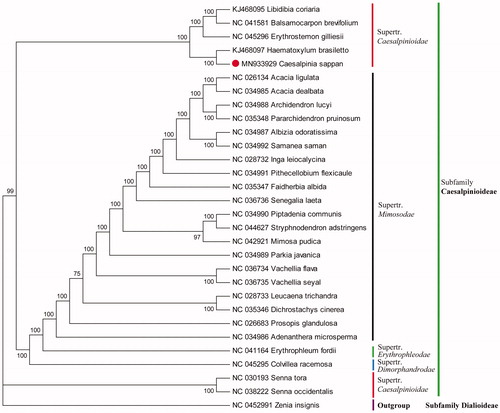
The complete cp genome of C. sappan was 160,192 bp with the typical quadripartite structure of angiosperms, including a large single-copy region (LSC) of 89,726 bp, a small single-copy region (SSC) of 18,358 bp, and the inverted repeat region (IR) of 26,054 bp. The genome harbored 127 genes, including 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content in the cp genome of C. sappan was 36.0%. To investigate the phylogenetic relationship of C. sappan, a total of 29 complete cp genome sequences of related species were downloaded from the NCBI database. Genome alignment was performed by MAFFT v7.307 (Katoh and Standley Citation2013). RAxML (Stamatakis Citation2014) was used to reconstruct a Maximum-Likelihood (ML) phylogenetic tree, with Rosa berberifolia (NC_045126) as outgroup (). The ML tree showed that C. sappan was closely related to Haematoxylum brasiletto, which formed an independent clade of the subfamily Caesalpinioideae (). The phylogenetic relationship of C. sappaan L. with genomic data was uncovered for the first time, largely enriching genetic resources for resolving the complex phylogeny relationship of Leguminosae.
Disclosure statement
The authors are grateful to the published genome data in the public database. No potential conflicts of interest and would be responsible for the content were declared by the authors.
Additional information
Funding
References
- Cai CQ, Zhao MB, Tang L, Tu PF. 2012. Study on chemical constituents from heartwood of Caesalpinia sappan. Chinese Tradit Herb Drugs. 2(43):230–233.
- Flora of China Editorial Committee of Chinese Academy of Sciences. 2010. Flora of China. Vol. 10. Beijing: Science Press & Missouri Botanical Garden Press; p. 1735.
- Gagnon E, Lewis GP, Solange Sotuyo J, Hughes CE, Bruneau A. 2013. A molecular phylogeny of Caesalpinia sensu lato: increased sampling reveals new insights and more genera than expected. S Afr J Bot. 89(89):111–127.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. doi.org/10.1101/256479
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7 improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Michael T, Pascal L, Tommaso P, U E S, Axel F, Ralph B, Stephan G. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Mueller M, Weinmann D, Toegel S, Holzer W, Unger FM, Viernstein H. 2016. Compounds from Caesalpinia sappan with anti-inflammatory properties in macrophages and chondrocytes. Food Funct. 7(3):1671–1679.
- Nirmal NP, Rajput MS, Prasad RGSV, Ahmad M. 2015. Brazilin from Caesalpinia sappan heartwood and its pharmacological activities: a review. Asian Pac J Trop Med. 6(8):5–14.
- Park J, Choi YG, Yun N, Xi H, Min J, Kim Y, Oh S. 2019. The complete chloroplast genome sequence of Viburnum erosum (Adoxaceae). Mitochondr DNA B. 4(2):3278–3279.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.