Abstract
The whole chloroplast genome sequence of Populus wulianensis was assembled and characterized. Its length was 156,553 bp, containing a large fragment of 84,726 bp and a small fragment of 16,551 bp separated by a pair of inverted repeat regions of 27,638 bp. A total of 129 genes were determined in the chloroplast genome, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The maximum-likelihood phylogenetic analysis showed that P. wulianensis was close to P. adenopoda but far from P. davidiana. The report here clearly displayed its origin and taxonomic status in Populus.
Populus wulianensis is mainly distributed in the hinterland of Kunyu and Wulian mountain in Shandong province of China (Fang et al. Citation1999). It was recognized as an important species in genus Populus since its special morphological feature and high resistance to drought and poor nutrients environment. A morphological analysis showed P. wulianensis as a hybrid between P. davidiana and P. adenopod possibly (Liang and Li Citation1986), with some characters same as P. davidiana and some others as P. adenopod. The genetic assays were also conducted to examine the taxonomic status of P. wulianensis with five barcoding DNAs (one ITS and four the complete chloroplast genome DNA fragments) and 14 nuclear SSR primers. The results showed the individuals of P. wulianensis really belong to the P. adenopod group without distinct introgression from P. davidiana (Zhang et al. Citation2018). In the present study, we suppose to sequence and characterize the cp genome of P. wulianensis to provide more evidence of its origin and phylogenetic status in Populus.
The fresh leaves of P. wulianensis were collected in Shandong Provincial Center of Forest Tree Germplasm Resources (36°37′N, 117°9′E). The voucher specimens were deposited in National Plant Specimen Resource Center and the manuscript of the specimen accession number is BNU 0034846. Total genomic DNA was extracted with the plant DNA extraction kit (TIANGEN, Beijing, China) by following the manual. Paired end (PE) reads were constructed according to the Illumina library preparation protocol and sequenced in Illumina HiSeq 2000 platform (Illumina, San Diego, CA). The cp genome of P. wulianensis was assembled by using the MITObim 1.8 (Hahn et al. Citation2013), and annotated by using the DOGMA. A neighbor-joining (NJ) tree with 1000 bootstrap replicates was inferred by using TreeBeST 1.9.29 (Vilella et al. Citation2009).
The assembled result showed that the cp genome of P. wulianensis (GenBank accession # MN829874) is 156,553 bp long, with the overall GC content of 33.2%. It is composed of a large single-copy (LSC) region of 84,726 bp and a small single-copy (SSC) region of 16,551 bp, which were separated by a pair of inverted repeat regions (IRa, b) of 27,638 bp. One hundred and twenty-nine genes were determined in the cp genome. A total of 110 genes occurred in a single copy but 19 genes were duplicated in the IR regions, including seven protein-coding genes, seven tRNA genes, and four rRNA genes.
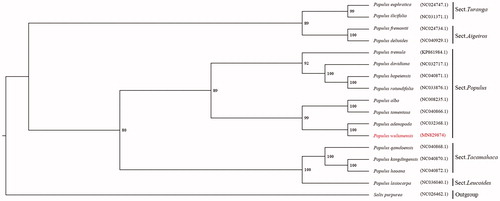
The maximum-likelihood phylogenetic tree was constructed with the cp genomes of 16 Populus species and one Salix species (as outgroup) based on the sequence alignments by MAFFT v7.307 (Katoh and Standley Citation2013) and GTRGAMMA model of RAxML (Silvestro and Michalak Citation2012) (). All the Populus species were divided into five sections (Sect. Turanga, Sect. Aigeiros, Sect. Populus, Sect. Tacamahaca, and Sect. Leucoides). Populus wulianensis was mostly close with P. adenopoda but far from P. davidiana in Sect. Populus. Thus, the study here clearly confirmed P. adenopoda as its possible mother parent through the cp genome evaluation, which could be significant for P. wulianensis using in future breeding programs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Fang ZF, Zhao SD, Skvortsov AK. 1999. Salicaceae. In: Wu Z, Raven P, editors. Flora of China (English version). Volume 4. Beijing, China: Science Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liang SB, Li XW. 1986. A new species of Populus from Shandong. Bull Bot Res. 6:137–139.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19(2):327–335.
- Zhang L, Wang MC, Ma T, Liu J. 2018. Taxonomic status of Populus wulianensis and P. ningshanica (Salicaceae). PhytoKeys. 108:117–129.