Abstract
Senna tora has been used as a famous traditional medicine both in China and India to treat skin diseases, cough, hepatitis, fever, and hemorrhoids. Here, the complete chloroplast (cp) genome sequence of S. tora was reported and characterized for the first time. The cp genome is 162,426 bp in length, with a large single copy (LSC) region of 90,673 bp and a small single copy (SSC) region of 18,001 bp separated by a pair of 26,791 bp inverted repeat (IR) regions. There were 130 predicted genes including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes in the genome, and the overall GC content of the genome is 36%. The phylogenetic analysis based on the cp genome data indicated that S. tora was closer to S. occidentalis.
Senna tora (L.) Roxb. (Fabaceae), an annual herbaceous undershrub widespread in tropical countries, has been used as a famous traditional medicine both in China and India to treat skin diseases, cough, hepatitis, fever, leprosy, dyspepsia, and hemorrhoids (Shakywar et al. Citation2011; Puri Citation2018). The seeds of C. tora were also commonly used as laxative and tonic in Asia (Tzeng et al. Citation2013; Lee et al. Citation2018). Besides, the seed extracts have been used as eco-friendly natural dyes for cotton and silk (Lee and Kim Citation2004). To provide genomic resources for investigating the evolution of S. tora, we reported the complete chloroplast (cp) genome sequences of this species and reveal its phylogenetic relationships within Caesalpinioideae.
Fresh leaves of S. tora were collected from Longping town of Jianshi county, Hubei, China (N30°48′24″, E110°1′47″, 1, 750 m). The voucher specimen (HSN12413) was deposited in the herbarium of South-Central University for Nationalities (HSN). The total genomic DNA was extracted and sequenced on Illumina HiSeq 4000 Platform at the Beijing Novogene Bioinformatics Technology Co., Ltd. (Nanjing, China). Raw data were used to de novo assemble the complete cp genome using SPAdes (Bankevich et al. Citation2012). The complete genome sequences were annotated using PGA (Qu et al. Citation2019) with manual adjustments.
The complete cp genome of S. tora (GenBank accession is MN971593) exhibits a typical quadripartite circular molecule with 162,426 bp in size. It is composed by a small single copy (SSC) region of 18,001 bp, a large single copy (LSC) region of 90,673 bp and a pair of inverted repeat (IR) regions of 26,791 bp. The GC content of this genome was 36.0%. The cp genome encodes an identical set of 113 predicted functional genes including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Most of the genes occurred in a single copy, while 17 genes (four rRNA genes, seven tRNA genes, and six protein-coding genes) occurred in double. Among the 113 genes, 15 of them had one intron, and three had two introns (clpP, rps12, and ycf3).
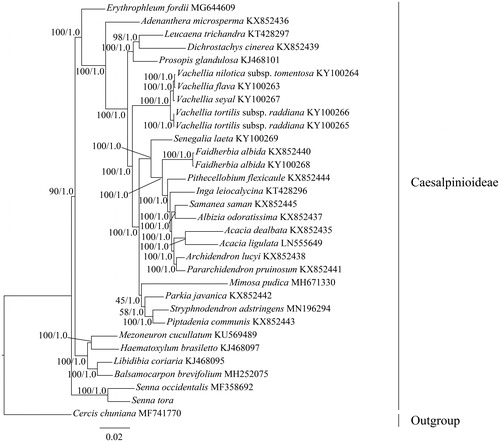
To confirm the phylogenetic position of S. tora, phylogenetic analysis was conducted based on the complete cp genome of this species and other 28 Caesalpinioideae species (including subspecies). Cercis chuniana belonging to Cercidoideae was used for outgroup. The sequences were aligned with MAFFT (Katoh and Standley Citation2013). The maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees were reconstructed using RAxML (Stamatakis Citation2014) and MrBayes (Ronquist et al. Citation2012). The ML and BI analyses generated the same tree topology (). The phylogenetic tree showed that the genus Senna was sister to the rest species of Caesalpinioideae and S. tora was closely related to S. occidentalis ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee GY, Cho B, Shin JY, Jang SI, Cho IS, Kim HY, Park JS, Cho CW, Kang JS, Kim JH, et al. 2018. Tyrosinase inhibitory components from the seeds of Cassia tora. Arch Pharm Res. 41(5):490–496.
- Lee YH, Kim HD. 2004. Dyeing properties and colour fastness of cotton and silk fabrics dyed with Cassia tora L. extract. Fibers Polym. 5(4):303–308.
- Puri BK. 2018. Editorial: the potential medicinal uses of Cassia tora Linn leaf and seed extracts. Rev Recent Clin Trials. 13(1):3–4.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shakywar Y, Jain A, Verma M, Panwar AS, Agarwal A. 2011. Pharmacognostical properties and their traditional uses of Cassia tora. Int J Pharm Biol Arch. 2(5):1311–1318.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tzeng T-F, Lu H-J, Liou S-S, Chang CJ, Liu I-M. 2013. Reduction of lipid accumulation in white adipose tissues by Cassia tora (Leguminosae) seed extract is associated with AMPK activation. Food Chem. 136(2):1086–1094.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.