Abstract
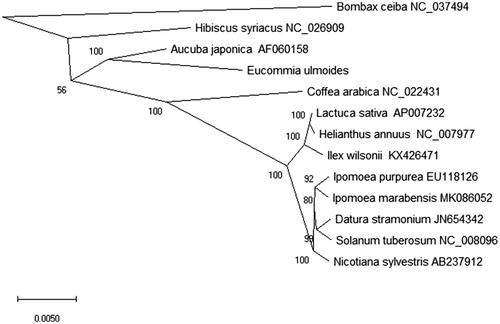
Eucommia ulmoides is an important traditional medicinal plant and as a common herb for orthopedic treatment in China. In this study, we reported and analyzed the complete chloroplast genome of E. ulmoides. The chloroplast genome length of E. ulmoides was 163,586 bp in size that has the typical quadripartite structure. It contained a large single-copy (LSC) region of 86,773 bp, a small single-copy (SSC) region of 14,167 bp and separated by two inverted repeat (IRa and IRb) regions of 31,323 bp. The overall GC content of the circular genome was 38.4% while A of 30.8% (50,496 bp), T of 30.8% (50,414 bp), C of 19.2% (31,341 bp), G of 19.2% (31,335 bp). In the chloroplast genome, 135 genes had been found, which included 89 protein-coding (PC) genes, 38 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. Phylogenetic relationship analysis shown that Eucommia ulmoides was clustered with Aucuba japonica in the evolutionary relationship by the Maximum-Likelihood (ML) method.
Eucommia ulmoides is the single extant species in the genus Eucommia, which also has shown a variety of important pharmacological activities and medicinal applications in China (Wang et al. Citation2014). It is native to China, which has been widely cultivated in central and southeastern regions at China and other countries in the world (Li et al. Citation2014). E. ulmoides as a traditional Chinese medicine can use for orthopedic treatment, whichi alao has several pharmacological properties such as antioxidant, anti-inflammatory, antiallergic, antimicrobial, anticancer, antiaging, cardioprotective and neuroprotective properties at the same time (Hussain et al. Citation2016). In summary, we reported and analyzed the chloroplast genome of E. ulmoides, we can expect that these data will be valuable for further studies of E. ulmoides in the evolutionary and phylogenetic relationship, more will conducive to the application of this traditional Chinese medicine in China.
Total genomic DNA was extracted from rind of Eucommia ulmoides that was collected from the herb market near Zhejiang Chinese Medical University at Hangzhou, Zhejiang, China (119.89E, 30.09 N). The genomic DNA was extracted using the modified CTAB method and stored at Zhejiang Chinese Medical University (No. ZJCMU-005), which was purified and sequenced. For quality control, we used the FastQC software (Andrews Citation2015) to remove the low-quality reads. We used the MitoZ software (Meng et al. Citation2019) to assemble and annotate the chloroplast genome of E. ulmoides. We had been drawn the physical map of the chloroplast genome using the OGDRAW software (Greiner et al. Citation2019). The chloroplast genome sequence had been deposited into NCBI GenBank (NCBI Accession No. MN0448772).
The chloroplast genome length of Eucommia ulmoides was 163,586 bp in size that has the typical quadripartite structure. It contained a large single-copy (LSC) region of 86,773 bp, a small single-copy (SSC) region of 14,167 bp and separated by two inverted repeat (IRa and IRb) regions of 31,323 bp. The chloroplast genome of E. ulmoides contained 135 genes, which included 89 protein-coding (PC) genes, 38 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. In one of IR region, 19 genes was found that contained 8 PCGs species, 7 tRNA genes species and 4 rRNA genes species. The overall GC content of the circular genome is 38.4% while A of 30.8% (50,496 bp), T of 30.8% (50,414 bp), C of 19.2% (31,341 bp), G of 19.2% (31,335 bp).
To stduy the phylogenetic and evolutionary relationship of Eucommia ulmoides, based on the chloroplast genomes of 13 plant species, we constructed the phylogenetic tree by the Maximum-Likelihood (ML) method. We used the MEGA X software (Kumar et al. Citation2018) to analysis the phylogenetic tree and performed using 2000 bootstrap values replicate for all the node by the ML methods. In the end, we used the iTOL version 4.0 (Letunic and Bork Citation2016) to edit the ML phylogenetic tree. Phylogenetic relationship analysis shown that Eucommia ulmoides was clustered with Aucuba japonica in the evolutionary relationship (). This research will be useful for phylogenetic and evolutionary studies for traditional medicinal plant.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Hussain T, Tan B, Liu G, Oladele OA, Rahu N, Tossou MC, Yin Y. 2016. Health-promoting properties of Eucommia ulmoides: a review. Evid Based Complement Alt Med. 2016(12):1–9.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v4: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Li Y, Wei YC, Li ZQ, Wang SH, Chang L. 2014. Relationship between progeny growth performance and molecular marker-based genetic distances in Eucommia ulmoides parental genotypes. Genet Mol Res. 13(3):4736–4746.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Research. 47(11):e63–e63.
- Wang DW, Li Y, Li L, Wei YC, Li ZQ. 2014. The first genetic linkage map of Eucommia ulmoides. J Genet. 93(1):13–20.