Abstract
Euodia ruticarpa var. bodinieri is a medicinal plant recorded in Chinese Pharmacopeia. Here, we report the complete chloroplast (cp) genome sequence of Euodia ruticarpa var. bodinieri. The cp genome is 158,745 bp in size and includes two inverted repeat regions of 27,101bp each, which is separated by a large single-copy region of 86,299 bp and a small single copy region of 18,244 bp. A total of 132 genes were predicted, including 37 tRNA, 8 rRNA, and 87 protein-coding genes. Phylogenetic analysis placed Euodia ruticarpa var. bodinieri under the family Rutaceae.
Euodia ruticarpa var. bodinieri (synonym: Tetradium ruticarpum) is one of the well-known medicinal herbs in China. The dried unripe fruit of E. ruticarpa var. bodinieri, known as ‘Wuzhuyu’, is recorded in Chinese Pharmacopeia, and it is also traditionally and ethnically used as a medicinal herb in Japan and Korea (Zhao et al. Citation2019). In traditional Chinese medicine (TCM), Wuzhuyu could be used either alone or in combination with other herbal medicines to cure headache, epigastric pain, menorrhalgia, dermatophytosis, celialgia, emesis and aphtha. In vitro metabolites of evodiamine with LC-MS/MS were elucidated (Zhou et al. Citation2010) and CYP isoforms involved in the metabolism of evodiamine were identified (Sun et al. Citation2013). However, the information of chloroplast (cp) genome of E. ruticarpa var. bodinieri is limited.
In this study, we aimed to characterize the complete cp genome sequence of E. ruticarpa var. bodinieri to serve as a valuable genomic resource. Total genomic DNA was extracted from the fresh leaves of E. ruticarpa var. bodinieri (Mcpherson et al. Citation2013), planted in Botanical Garden, Institute of Chinese Materia Medica, Hunan Academy of Chinese Medicine (N28°13′28.15″, E112°56′26.96″). Additional leaf specimens were kept in Hunan Herbarium of Chinese Traditional Medicine under the collection number HUTM100004.
The genome was sequenced using a combination of PacBio RS and Illumina sequencing platforms. The Illumina data were used to evaluate the complexity of the genome and correct the PacBio long reads. The output was a 3 Gb raw data of paired-end reads, further assembled using ABySS, canu and GapCloser software (Li et al. Citation2019). Ab initio prediction method was used to get gene models, which were identified using GeneMark (Besemer and Borodovsky Citation2005). All gene models were blastp against non-redundant (NR in NCBI) database, SwissProt, KEGG, and COG to perform functional annotation using blastp module (Rahman et al. Citation2013). Then, tRNA were identified using the tRNAscan-SE and rRNA were determined using the RNAmmer (Malhotra et al. Citation2012) .
The complete cp genome of E. ruticarpa var. bodinieri (GenBank accession number: MT134114) is 158,745 bp in length, displaying a quadripartite structure that contains a pair of inverted repeats (IR) regions (27,101 bp each), separated by a large single-copy (LSC) region (86,299 bp) and a small single-copy (SSC) region (18,244 bp). There are 132 genes reported, including 8 rRNA genes, 37 rRNA genes, and 87 protein-coding genes. The overall GC content of the cp genome was 38.336%.
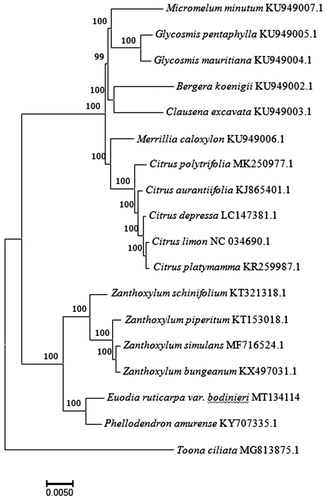
A maximum-likelihood (ML) tree was constructed with 1000 bootstrap replicates using FastTree software for phylogenetic analysis (Liu et al. Citation2011). A subset of another 16 species from the family Rutaceae was included, with Toona ciliata from Meliaceae as outgroup. The ML analysis showed that E. ruticarpa var. bodinieri is placed under the family Rutaceae (). The taxonomic status of E. ruticarpa var. bodinieri exhibits a closest relationship with Phellodendron amurense. This finding could serve as a valuable genomic resource providing insight into this medicinal plant species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Besemer J, Borodovsky MJNar. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33(Web Server):W451–W454.
- Li Y, Tang B-L, Ren X-B, Dang Y-R, Sun L-L, Zhang X-Y, Chen X-L, Qin Q-L, Wang P. 2019. Complete genome sequence of Flavobacterium arcticum SM1502T, exhibiting adaption to the Arctic marine salty environment. Mar Genomics. 47:100670.
- Liu K, Randal LC, Tandy W, Rongling W. 2011. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS One. 6(11):e27731.
- Malhotra J, Dua A, Saxena A, Sangwan N, Mukherjee U, Pandey N, Rajagopal R, Khurana P, Khurana JP, Lal R. 2012. Genome sequence of Acinetobacter sp. strain HA, isolated from the gut of the polyphagous insect pest Helicoverpa armigera. J Bacteriol. 194(18):5156–5156.
- Mcpherson H, Merwe MVD, Delaney SK, Edwards MA, Henry RJ, Mcintosh E, Rymer PD, Milner ML, Siow J, Rossetto MJBE. 2013. Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol. 13(1):8.
- Rahman AYA, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, Hou S, Ong SY, Ng FL, Lee LS, et al. 2013. Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics. 14(1):75.,
- Sun HZ, Fang ZZ, Cao YF, Sun XY, Hong MJPR. 2013. Investigation of the in vitro metabolism of evodiamine: characterization of metabolites and involved cytochrome p450 isoforms. Phytother Res. 27(5):705–712.
- Zhao Z, He X, Han W, Chen X, Liu P, Zhao X, Wang X, Zhang L, Wu S, Zheng XJJoe. 2019. Genus Tetradium L.: a comprehensive review on traditional uses, phytochemistry, and pharmacological activities. J. Ethnopharmacol. 231:337–354.
- Zhou X, Zhao Y, Lei P, Cai Z, Liu HJJoss. 2010. Chromatographic fingerprint study on Evodia rutaecarpa (Juss.) Benth by HPLC/DAD/ESI‐MSn technique. J Sep Science. 33(15):2258–2265.