Abstract
The complete mitochondrial genome (mitogenome) of diamond dove (Geopelia cuneata) was first determined in this study. The mitogenome is 17,880 bp in size and composed of 22 transfer RNA genes, 13 protein-coding genes, two ribosomal RNA genes, and a control region. The overall nucleotide composition is 30.11% of A, 13.66% of G, 25.11% of T, and 31.13% of C. The mitochondrial structure and gene order were similar to other Columbidae species. Phylogenetic analysis suggested that G. cuneata was placed as sister to G. striata of the same genus. The data could provide valuable information for further studies on conservation genetics and evolution of G. cuneata.
Diamond dove (Geopelia cuneata) is a small species belonging to the family Columbidae (BirdLife International Citation2016). This species is native to inland wetlands and hot deserts of Australia, and introduced to Puerto Rico (Schleucher Citation1993; BirdLife Inernational Citation2016). It is popular as a pet or an ornamental bird due to its colorful appearance (BirdLife Inernational Citation2016). However, little is known about the genetic background of this economic animal. Here, we describe the complete mitochondrial genome (mitogenome) of G. cuneata.
A captive-bred specimen of G. cuneata was collected from Nanjing Pet Market (N32°01′, E118°84′) and stored in College of Biology and the Environment, Nanjing Forestry University (Accession number: QQW202001). Genomic DNA was extracted using the Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China). Polymerase chain reaction was carried out using the Taq PCR Master Mix (Sangon Biotech, Shanghai, China). Sanger sequencing was performed by Sangon Biotech (Shanghai, China). The complete mitogenome of G. cuneata is a typical circular DNA molecule with 17,880 bp in size (GenBank accession number: MN930521). Its overall nucleotide composition is 30.11% of A, 13.66% of G, 25.11% of T, and 31.13% of C. There are 22 transfer RNA genes, 13 protein-coding genes (PCGs), 2 ribosomal RNA genes and a control region in the mitogenome. Almost all the PCGs use ATN as the initiation codon except for COI starting with GTG. Ten genes (ATP6, ATP8, COI, COII, Cyt b, ND1, ND3, ND4L, ND5, ND6) end with the typical stop codon TAA, TAG, AGA, or AGG, whereas the remaining three genes (COIII, ND2, and ND4) used the incomplete T. The gene arrangement in G. cuneata mitogenome was identical to other Columbidae species (Zhang et al. Citation2015; Huang et al. Citation2016; Soares et al. Citation2016; Bruxaux et al. Citation2018).
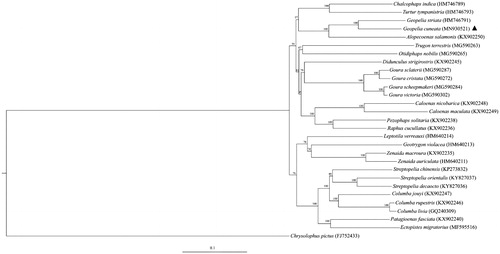
Phylogenetic analysis of Columbidae species was performed based on the concatenated datasets of 13 PCGs using the maximum-likelihood method in IQ-tree (Nguyen et al. Citation2015). Phylogenetic tree suggested that G. cuneata was placed as sister to G. striata of the same genus (). Moreover, the conservative relationships between Columbidae species were revealed by the ML tree, as the species from the same genera were initially clustered together.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- BirdLife International. 2016. Geopelia cuneata. The IUCN Red List of Threatened Species 2016: e.T22690705A93284384. [accessed 2020 Mar 9]. https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T22690705A93284384.en
- Bruxaux J, Gabrielli M, Ashari H, Prŷs-Jones R, Joseph L, Milá B, Besnard G, Thébaud C. 2018. Recovering the evolutionary history of crowned pigeons (Columbidae: Goura): implications for the biogeography and conservation of New Guinean lowland birds. Mol Phylogenet E. 120:248–258.
- Huang ZH, Tu FY, Murphy RW. 2016. Analysis of the complete mitogenome of Oriental turtle dove (Streptopelia orientalis) and implications for species divergence. Biochem Syst Ecol. 65:209–213.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol E. 32(1):268–274.
- Schleucher E. 1993. Life in extreme dryness and heat: a telemetric study of the behavior of the diamond dove Geopelia cuneata in its natural habitat. Emu. 93(4):251–258.
- Soares AE, Novak BJ, Haile J, Heupink TH, Fjeldså J, Gilbert MT, Poinar H, Church GM, Shapiro B. 2016. Complete mitochondrial genomes of living and extinct pigeons revise the timing of the columbiform radiation. BMC Evol Biol. 16(1):230.
- Zhang RH, He WX, Xu T. 2015. Characterization of the complete mitochondrial genome of the king pigeon (Columba livia breed king). Mitochondrial DNA. 26(3):491–492.