Abstract
The complete mitochondrial genome of a nursery-web spider Dolomedes angustivirgatus is 14,783 bp in length and contains a standard set of 37 genes including 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs and a putative control region. The A + T content of the major strand is 77.0%. All the protein-coding genes are initiated by typical ATN codons, except for COII and COIII, which use a special start codon TTG. The truncated stop codon (T) occurs in ND4L, whereas the rest 12 genes end with the canonical stop codon (TAA and TAG). 15 tRNAs cannot be folded into typical cloverleaf-shaped secondary structure. The control region is 1084 bp in length and contains a long tandem repeat region. The result of phylogenetic analysis show that D. angustivirgatus is nested within Lycosoidea and recovered as sister to the cluster of three wolf spiders (Lycosidae).
Mitochondrial genome has been wildly used as an effective molecular marker for studying arachnid phylogeny (Fahrein et al. Citation2007). Spiders (order Araneae) are the second-largest arachnid groups, but there is a lack of sufficient mitochondrial genome sequence data for many taxa. In this study, we determined the complete mitochondrial genome of a nursery-web spider Dolomedes angustivirgatus, which is the first mitochondrial genome sequenced to date in the family Pisauridae.
The sampled specimen was collected from a rice paddy in Yuyao (E121°09′, N30°03′), Zhejiang Province, China, and deposited in the insect specimen room of China Jiliang University, Hangzhou, China, with an accession number CJLU-YY-2019018. Total genomic DNA was extracted from leg tissue of a spider individual using the DNeasy Tissue Kit (Qiagen, Hilden, Germany). The complete mitochondrial genome sequence was amplified by over-lapping extension PCR, according to the method described previously (Wang et al. Citation2016). Maximum likelihood (ML) was used to reconstruct a mitogenomic phylogeny of Araneae, including 33 spiders and a horseshoe crab Limulus polyphemus as an outgroup (Lavrov et al. Citation2000; Guindon and Gascuel Citation2003).
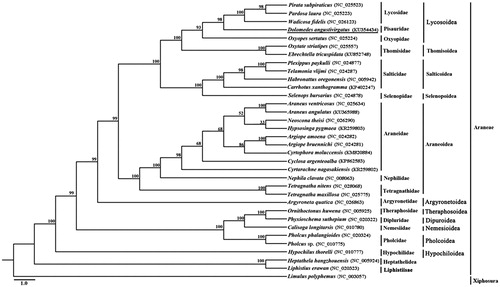
The complete mitochondrial genome of D. angustivirgatus (GenBank accession number: KU354434) is a typical circular DNA with the length of 14,783 bp. The nucleotide composition is significantly A + T biased, as the overall A + T content of the major strand accounts for 77.0%. Within the complete mitochondrial genome, a standard set of 37 genes was identified, including 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a putative control region (CR). The gene arrangement is found to be identical to most mitochondrial genomes of Entelegynae spiders. All the PCGs are initiated by typical ATN codons, except for COII and COIII, which use a special start codon TTG. 12 PCGs are terminated by the canonical stop codon (TAA and TAG), whereas ND4L ends with an incomplete stop codon (T). Among 22 tRNAs, only seven tRNAs (tRNACys, tRNAGln, tRNAGly, tRNAIle, tRNALeu(CUN), tRNAPro and tRNATyr) can be folded into typical cloverleaf-shaped secondary structure. Three tRNAs (tRNASer(AGN), tRNASer(UCN) and tRNAAla) lack the dihydrouracil (DHU) arm and the rest 12 tRNAs cannot form the TΨC arm. The control region is 1084 bp in length and contains a long tandem repeat region, comprising three full 266 bp copies. As shown in both ML tree, D. angustivirgatus is nested within Lycosoidea and recovered as sister to the cluster of three wolf spiders (Lycosidae) (). Our data will be provide fundamental data for further phylogenetic and population genetic studies of the Araneae.
Figure 1. Phylogenetic reconstruction of 33 spider species based on nucleotide sequences of mitochondrial 13 protein-coding genes using Maximum likelihood (ML). Numbers at the branch indicate the percentages from ML bootstrapping. GenBank accession number of a given mitochondrial genome is parenthesized following the name of each spider species. Spider determined in this study is underlined.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Fahrein K, Talarico G, Braband A, Podsiadlowski L. 2007. The complete mitochondrial genome of Pseudocellus pearsei (Chelicerata: Ricinulei) and a comparison of mitochondrial gene rearrangements in Arachnida. BMC Genomics. 8(1):386.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Boil. 52(5):696–704.
- Lavrov DV, Boore JL, Brown WM. 2000. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol Evol. 17(5):813–824.
- Wang ZL, Li C, Fang WY, Yu XP. 2016. The complete mitochondrial genome of two Tetragnatha spiders (Araneae: Tetragnathidae): severe truncation of tRNAs and novel gene rearrangements in Araneae. Int J Biol Sci. 12(1):109–119.
