Abstract
The complete mitochondrial genome of smallscale blackfish, Girella melanichthys (Perciformes: Kyphosidae), was analyzed using MGISEQ-2000 platform. The assembled mitogenome had 16,518 nucleotides (overall A + T contents 56.2%), containing canonical 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 putative control region. Among 13 PCGs, seven genes (ND2, ND3, ND4, COII, COIII, CytB, ATPase6) had incomplete stop codons. The phylogenetic study revealed that G. melanichthys is more closely related to G. leonina than G. punctate. This study will be useful to shed light on the evolution of the family Kyphosidae and the genus Girella.
The Kyphosidae has been considered a group that are taxonomically difficult for a long time (Humann Citation1994). Kyphosid species are found in temperate, subtropical, and tropical waters in the Indo-Pacific (Sakai and Nakabo Citation2004) and the Atlantic (Smith-Vaniz et al. Citation1999) regions. In East Asia, the genus Girella is represented by three species, Girella punctata, G. leonina and G. mezina (Itoi et al. Citation2007). Addition to this, smallscale blackfish Girella melanichthys is commonly inhabited in the coast of Jeju Island in Korea. Since smallscale blackfish has a very similar shape to largescale blackfish (Girella punctata), not only biological examines but also genetic studies on smallscale blackfish are sufficiently required. The aim of this study is to provide mitochondrial genome information of smallscale blackfish through NGS analysis tool.
The specimen was caught in the Jeju coast in Korea (33°15′38.8″ 126°42′39.1″E). The voucher specimen (Sample no. MFDS-FDO10) was deposited at the Department of Food Engineering, Pukyong National University, under the funding from Ministry of Food and Drug Safety. DNA library was constructed using MGIEasy DNA Library Prep Kit (MGI, China) and sequenced by MGI MGISEQ-2000 with 150 paired-ends. The obtained raw reads were cleaned with Cutadapt 1.9 (Martin Citation2011) and the cleaned data were mapped to previously reported mitogenome of G. punctata (Accession No. NC013137) using Geneious 11.1.3 (Kearse et al. Citation2012). The pipeline of MitoFish (Sato et al. Citation2018) was used for annotation step and phylogenetic analysis was processed with MEGA X (Kumar et al. Citation2018).
The assembled mitogenome had 16,518 nucleotides (overall A + T contents 56.2%), containing canonical 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 putative control region. Among 13 PCGs, seven genes (ATPase6, COII, COIII, CytB, ND2, ND3, ND4) included incomplete stop codons. In case of transfer RNA, uncommon codons were found in Leucine (UAA, UAG), Histidine (GCU), and Serine (UGA). The ND6 gene and 8 tRNA (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro) genes were positioned on the H-strand. All these information and assembled sequence were submitted to the GenBank (Accession No. MT084559).
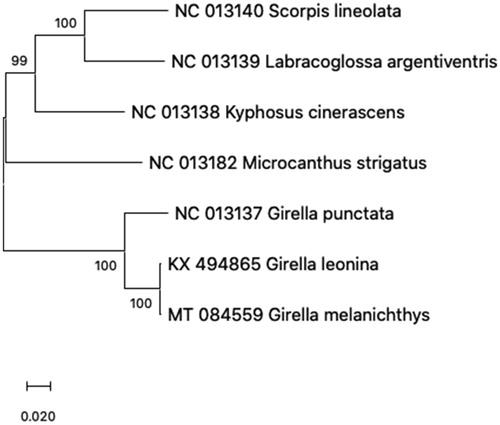
The phylogenetic analysis with currently reported 6 mitogenome sequences in Kyphosidae showed that G. melanichthys is more close to the G. leonina (Accession No. KX494865) than G. punctata (Accession No. NC013137) (). This study will be useful to understand evolutionary and phylogenetic exploration of the family Kyphosidae.
Figure 1. Phylogenetic tree of 7 species in family Kyphosidae. The 6 mitogenome sequences from GenBank database and Girella melanichthys mitogenome sequence were aligned using ClustalW and phylogenetic analysis was conducted by Maximum Likelihood method with 1000 bootstrap. The percentage at each node is the bootstrap probability.
Acknowledgements
Special thanks to Dr. Moo-Sang Kim and the MOAGEN Corporation for their supporting to analyze NGS data.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Humann P. 1994. Reef fish identification Florida-Caribbean-Bahamas. 2nd ed. Jacksonville, FL: New World Publications.
- Itoi S, Saito T, Washio S, Shimojo M, Takai N, Yoshihara K, Sugita H. 2007. Speciation of two sympatric coastal fish species, Girella punctata and Girella leonina (Perciformes, Kyphosidae). Org Divers Evol. 7(1):12–19.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Bio Evol. 35(6):1547–1549.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12.
- Sakai K, Nakabo T. 2004. Two new species of Kyphosus (Kyphosidae) and a taxonomic review of Kyphosus bigibbus Lacepède from the Indo-Pacific. J Appl Ichthyol. 51(1):20–32.
- Sato Y, Miya M, Fukunaga T, Sado T, Iwasaki W. 2018. MitoFish and MiFish Pipeline: a mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol Bio Evol. 35(6):1553–1555.
- Smith-Vaniz W, Collette B, Luckhurst B. 1999. Fishes of Bermuda: history, zoogeography, annotated checklist, and identification keys. American Society of Ichthyologists and Herpetologists, Special Publication. Vol. 4. https://pubs.er.usgs.gov/publication/70162457
