Abstract
Liquidambar acalycina is a popular tree species with high ornamental and economic values because of the leaf shape and coloration. In this study, chloroplast genome of L.acalycina was assembled using Illumina highlights pair-end sequencing data. The chloroplast genome has a quadripartite structure with 160,433 bp in length. It contains a pair of 26,274 bp inverted repeat (IR) regions, which were separated by large single-copy (LSC: 88,963bp) region and small single-copy (SSC: 18,922 bp) region. A total of 133 genes were annotated, including 88 protein-coding genes (PCGs), 37 tRNAs, and 8 rRNAs. The overall GC content was 36.1%. The phylogenetic analysis revealed that L. acalycina has a close relationship with L. fomosana. This complete chloroplast genome is of great value in the genetic research of L. acalycina.
Liquidambar acalycina Chang is a deciduous ornamental tree species in the genus Liquidambar, belong to the family Altingiaceae (formerly Hamameliaceae), native to subtropical areas of East Aisa, specifically in the Southern China (Margaret and Clifford Citation1994). The species is naturally distributed in the mountains above 600 m and often cultivated as an attractive ornamental tree for gardens worldwide, which is hardy down to –15 °C. The most distinctive character is the red flush of the leaves in spring and autumn. A particularly new plant variety has been named ‘Burgundy Flush’ (Hsu and Andrews Citation2005) and more under development. However, phenotypic characteristics of this species are very similar to L. formosana, especially for young plants. It is a big challenge for identification and certification of candidate varieties. Information of chloroplast genomes has been extensively applied for understanding plant genetic diversity and variety identification. In this study, we assembled the complete chloroplast genome of L. acalycina based on the whole-genome Illumina sequencing highlights simple dataset.
Fresh leaves of L. acalycina were collected from the top of wild seedlings (nearly 15 m high) in Hefeng County, Hubei Province, China (110°5′E, 29°48′N; asl:1206 m). The voucher specimen was deposited in the National Forest Genetic Resources Platform with the accession number 111C00003001000002 and DNA backups were deposited in the Chinese Academy of Forestry in Beijing (19°33′E, 33°15′N).The genomic DNA was extracted by the CTAB method (Doyle and Doyle Citation1987) and then sequenced by Illumina HiSeq X Ten (Majorbio, Shanghai, China). We used the software SOAP denovo v2.04 to assemble the chloroplast genome. The resulting contigs were linked based on overlapping regions after being aligned to L. formosana (NC_023092) (Dong et al. Citation2013). Blastp was used to annotate the highlighted data with manual adjustment. The annotated chloroplast genome of L. acalycina has been deposited into the GenBank with the accession number MT079212.The chloroplast genome has a circular quadripartite structure with 160,433 bp in length. It contained the large single-copy (LSC: 88,963 bp) region and small single-copy (SSC: 18,922 bp) region with a pair of inverted repeat (IR: 26,274 bp) regions. The plastid genome harbors 133 unique genes, including 88 protein-coding genes (PCGs), 37 tRNAs, and 8 rRNAs. The genomic GC content of this species was 36.1%. Among IR regions, four rRNAs genes, seven tRNAs genes, and eight protein-coding genes were found.
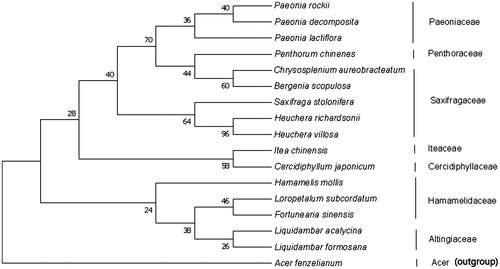
To explore the evolution status of L. acalycina, we inferred the phylogenetic relationships based on the complete chloroplast genomes of 18 species. The phylogenetic tree was constructed with RaxML based on 19 complete chloroplast genome sequences of Acer fenzelianum (NC_045527) as an outgroup (). All sequences were aligned using MAFFT (Nakamura et al. Citation2018) and using maximum-likelihood (ML) analysis based on the Neighbor-Join and BioNJ algorithms methods in MEGA 7 (Felsenstein Citation1985; Kumar et al. Citation2016). The phylogenetic analysis revealed that L. acalycina has close relationship with L. formosana. This complete chloroplast genome can be used for genetic studies and new plant varieties marker development of L. acalycina.
Figure 1. Molecular phylogenetic analysis by maximum-likelihood (ML) method based on the complete chloroplast genome sequences of L. acalycina and other 16 species. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. Their accession numbers are as follows: Liquidambar formosana: NC_023092; Bergenia scopulosa: NC_036061; Loropetalum subcordatum: NC_037694; Paeonia rockii: NC_037772; Hamamelis mollis: NC_037881; Saxifraga stolonifera: NC_037882; Itea chinensis: NC_037884; Cercidiphyllum japonicum: NC_037940;Paeonia decomposita: NC_039425; Chrysosplenium aureobracteatum: NC_039740; Paeonia lactiflora: NC_040983; Fortunearia sinensis: NC_041487; Heuchera richardsonii: NC_042923; Heuchera villosa:NC_042924; Penthorum chinense: NC_023086; Acer fenzelianum: NC_045527.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dong W, Xu C, Cheng T, Lin K, Zhou S. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of saxifragales. Genome Biol Evol. 5(5):989–997.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4):783–791.
- Hsu E, Andrews S. 2005. Tree of the year: Liquidambar. International Dendrology Society Yearbook. 2004:11-45.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Margaret TH, Clifford RP. 1994. Genetic divergence in Liquidambar styraciflua, L. formosana, and L. acalycina (Hamamelidaceae). Systematic Botany. 19:308–316.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
