Abstract
In this study, we sequenced the complete mitochondrial genome of the jellyfish Aurelia coerulea (A. coerulea). The mitochondrial genome was a linear form (16,748 bp long, 65.7% AT), consisting of 13 protein coding genes (PCGs) (cox1, cox2, atp8, atp6, cox3, nad2, nad5, nad6, nad3, nad4L, nad1, nad4, and cytB), 2 rRNAs (12S and 16S), and 2 tRNAs (trnM and trnW). Mitochondrial gene arrangement of the A. coerulea was identical when compared to already-known mitochondrial genomes of Aurelia species. The maximum-likelihood (ML) phylogenetic tree using of 13 mitochondrial proteins showed that A. coerulea collected from Korea waters was closely related to A. limbata.
Moon jellies of the genus Aurelia are widely distributed in the world’s oceans, and their outbreaks have been reported globally (Ki et al. Citation2008). All species in the genus are morphologically similar, making it difficult to distinguish between species. However, molecular studies indicate that there are at least 13 cryptic species in the genus (Dawson et al. Citation2005). Aurelia coerulea (A. coerulea or Aurelia sp.1) is a genetically and morphologically distinct form (Scorrano et al. Citation2017), and its occurrence has been reported from the Northwest Pacific oceans, the coasts of California, and the Mediterranean coasts, expanding geographic ranges (Dawson and Jacobs Citation2001; Ki et al. Citation2008; Scorrano et al. Citation2017). Thus, it is imperative to analyze the complete mitochondrial genome of the jellyfish to better understand the global distribution and the molecular phylogenetic relationship of the Aurelia. In the present study, we sequenced the complete mitochondrial genome of A. coerulea, and investigated their genomic structures and molecular phylogenetic relationships using mitochondrial proteins.
The specimens of A. coerulea were collected from Susan-port (38°04′47.2′′N, 128°40′27.9′′E) in Yangyang, Korea, on 9 September 2015. Total genomic DNA was extracted from the jellyfish umbrella by using the cetyl-trimethylammonium bromide method (Ausubel et al. Citation1989) and the remaining part of the specimen was stored in the Department of Biotechnology, Sangmyung University, Korea (Accession No. RH58). Next-generation sequencing (NGS) was subjected to the gDNA (Miseq, Illumina, San Diego, CA), and paired end reads of mitochondrial genome sequences were assembled and annotated using MITOS (Bernt et al. Citation2013) and Geneious version 9.1.3 (Geneious, Auckland, New Zealand), respectively. A phylogenetic tree was reconstructed by using concatenated amino acid sequences of 13 protein coding genes (PCGs) in MEGA X (Kumar et al. Citation2018).
The complete mitochondrial genome of A. coerulea was sequenced at 16,748 bp in length (GenBank No. MT023105; 29.46% A, 36.25% T, 16.36% G, 17.93% C), and had the second highest GC% (34.29%) in the available Aurelia genomes reported previously. The genome contains 13 PCGs (cox1, cox2, atp8, atp6, cox3, nad2, nad5, nad6, nad3, nad4L, nad1, nad4, and cytB), 2 rRNAs (12S, 16S rRNA), and 2 tRNAs (trnM, trnW). In addition, it has a putative control region between cytB and 16S rRNA. The 17 mitochondrial genes order is completely identical to those of other Aurelia species, such as A. aurita (DQ787873), A. limbata (MK527107), Aurelia sp.3 (LC005413), and Aurelia sp.4 (LC005414) (Karagozlu et al. Citation2019). Specifically, two genes cox1 and 16S rRNA encoded in minority strand and the rest 15 genes encoded in majority strand (Shao et al. Citation2006). Mitochondrial PCGs of A. coerulea has four different start codons (ATG, ATC, ATA, and ATT) and four stop codons which are TAG, TAA, incomplete T(AA).
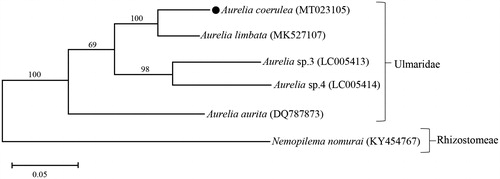
Molecular phylogenetic tree () using 13 PCGs of Aurelia showed that our A. coerulea formed a cluster with A. limbata which was collected from Yangyang City, Korea. These two species have been reported from Korean coastal waters, in which A. limbata only occurred northeastern coasts of Korea in cold seasons (Chang et al. Citation2016). This relationship was in accordance with previous molecular Aurelia phylogeny revealed by nuclear rDNA ITS-1 and mitochondrial cox1 markers (Dawson et al. Citation2005). These molecular approaches are very effective to discriminate cryptic species in the genus Aurelia (Ki et al. Citation2008), and to resolve deep phylogenetic relationships and ecological adaptations of the organisms (Ramšak et al. Citation2012). By considering cryptic Aurelia species, their mitochondrial genomes are still unknown, and further data should be recorded from the jellyfishes.
Figure 1. Molecular phylogeny of Aurelia coerulea in the family Ulmaridae. A relative jellyfish Nemopilema nomurai (Rhizostomeae) was included as the out-group. The phylogeny tree was reconstructed using concatenated amino acid sequences of 13 mitochondrial protein coding genes and the maximum-likelihood (ML) algorithm with the JTT matrix-based model in MEGA X software. Bootstrap proportions (BP) of the 1000 times replications were incorporated into the ML tree. The branch lengths are proportional to the scale given. Aurelia coerulea determined here represents with a black dot.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1989. Current protocols in molecular biology. New York (NY): John Wiley and Sons.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phyloenet Evol. 69(2):313–319.
- Chang SJ, Kim JN, Yoon WD, Ki JS. 2016. First record of two cold-water jellyfishes Aurelia limbata and Parumbrosa polylobata (Scyphozoa: Semaeostomeae: Ulmaridae) in Korean coastal waters. Anim Syst Evol Divers. 32(4):272–280.
- Dawson MN, Gupta AS, England MH. 2005. Coupled biophysical global ocean model and molecular genetic analyses identify multiple introductions of cryptogenic species. Proc Natl Acad Sci USA. 102(34):11968–11973.
- Dawson MN, Jacobs DK. 2001. Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa). Biol Bull. 200(1):92–96.
- Karagozlu MZ, Seo Y, Ki JS, Kim CB. 2019. The complete mitogenome of brownbranded moon jellyfish Aurelia limbata (Cnidaria, Semaeostomeae, Ulmaridae) with phylogenetic analysis. Mitochondrial DNA B. 4(1):1875–1876.
- Ki JS, Hwang DS, Shin K, Yoon WD, Lim D, Kang YS, Lee Y, Lee JS. 2008. Recent moon jelly (Aurelia sp.1) blooms in Korean coastal waters suggest global expansion: examples inferred from mitochondrial COI and nuclear ITS-5.8S rDNA sequences. ICES J Mar Sci. 65(3):443–452.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Ramšak A, Stopar K, Malej A. 2012. Comparative phylogeography of meroplanktonic species, Aurelia spp. and Rhizostoma pulmo (Cnidaria: Scyphozoa) in European Seas. Hydrobiologia. 690(1):69–80.
- Scorrano S, Aglieri G, Boero F, Dawson MN, Piraino S. 2017. Unmasking Aurelia species in the Mediterranean Sea: an integrative morphometric and molecular approach. Zool J Linnean Soc. 180:243–267.
- Shao Z, Graf S, Chaga O Y, Lavrov D V. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): A linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 381:92–101. doi:10.1016/j.gene.2006.06.021.
