Abstract
Economic value development of peony is limited by the fuzzy genetic relationships. Phylogeny investigation, genomic resource conservation, and genome evolution could be elucidated by the chloroplast genome (cp) on account of the extremely conservative structure. Paeonia lactiflora ‘Lv He’, a new peony variety blooming with green flowers, possess excellent ornamental value. Molecular structure and phylogenetic analysis of the plastome of Paeonia lactiflora ‘Lv He’ was resolved in this study. The complete cp genome contained a typical quadripartite structure with a length of 152, 702 bp and an overall 38.4% GC content. The circular molecule of the genome was constituted of a large single-copy (LSC) region of 84,373 bp with 36.7% GC content, a small single-copy (SSC) region of 16,969 bp with 32.7% GC content, and a pair of inverted repeat (IRa and IRb) regions of 25,680 bp each with 43.1% GC content. The genome annotated 115 genes, containg 81 protein-coding genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Phylogenetic analysis revealed a sister taxonomical relationship to P. lactiflora and P. obovate.
Paeonia lactiflora Pall belongs to Paeoniaceae is an important ornamental, pharmaceutical, and an immerging oil plant (Eason et al. Citation2002). Germplasm resources of P. lactiflora have changed greatly since the repeated hybridizations over thousands of years and the fluctuating habitats of wild plants which created affluent genetic diversity (Li et al. Citation2011). Appropriate molecular marker system has not been well established to date. It is necessary to find potential genetic markers suitable for identification, evaluation, preservation, innovation and utilization of P. lactiflora germplasm.
Complete cp genome which could increase phylogenetic resolution at a low taxonomic level has been widely used for phylogenetic studies (Yu et al. Citation2019). P. lactiflora ‘Lv He’ blooming with green flower is a peony variety generated by seedling selection from seeds introduced from Japan in 2006 (). The plant was conserved in National Peony Gene Bank of Luoyang, China (N34°42′29.70′′, E112°23′36.14′′). The specimen of ‘Lv He’ is stored in the Paeonia Specimen Museum of Henan University of Science and Technology under accession number PL2018041501. Cp genome was revealed to provide a basis for genetic resources developing and phylogenetic relationship clarifying with species in genus Paeonia.
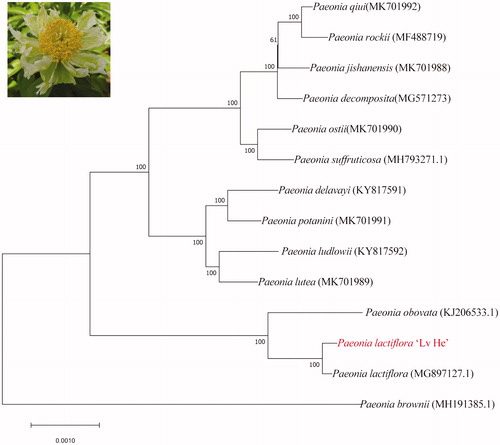
Figure 1. Phenotype and phylogenetic tree of Paeonia lactiflora ‘Lv He’. Maximum likelihood phylogenetic tree was constructed by clustering with the Paeoniaceae family including all the wild tree peony and three Paeonia lactiflora species. Numbers in the node are the bootstrap values from 1000 replicates.
Sample collection, genomic DNA extraction, library preparation and sequencing were performed according our previous studies (Guo et al. Citation2018). Sequences were assembled according to the SOAPdenovo-2.04 pipeline (Luo et al. Citation2012). SSPACE-3.0 and GapCloser-1.12 were devoted to the sequences extending and gaps filling (Acemel et al. Citation2016). The already published cp genome of Paeonia was used as reference. OGDRAWv1.3.1 (Greiner et al. Citation2019) was applied for annotation and mapping. tRNAscanSEv1.21 was used for tRNA prediction (Lowe and Eddy, Citation1997). The plastome was submitted to GenBank under the accession number of MN149612.
The plastome of Paeonia lactiflora ‘Lv He’ possessed a tetragonal structure with an entire length of 152,702 bp and an overall GC content of 38.4%. The plastid genome was composed of LSC region of 84,373 bp in length with 36.7% of GC content, SSC region of 16,969 bp having 32.7% of GC content, and 25,680 bp in each of two IR regions (IRa and IRb) with 43.1% of GC content. Altogether, 115 genes were annotated including 85 protein-coding genes, 30 tRNA genes, and four rRNA genes. Thereinto, 15 genes contain a single intron and three genes have a pair of introns. There are 22 two-copy genes located within IR regions.
Phylogenetic relationship was revealed by unrooted phylogenetic tree reconstruction utilizing Clustalw2 and MEGA5.2 (Tamura et al., Citation2011). Plastome of P. obovate, P. brownie, P. lactiflora and ten tree peony species were applied for phylogenomic analysis (). The result revealed that P. brownie was gathered into a single branch, while the rest taxa were clustered into one clade, which means P. lactiflora Pall has a closer relationship with P. suffruticosa rather than P. brownie. Subgroup I contained ten tree peony species, while Subgroup II was consisted by P. lactiflora ‘Lv He’, P. lactiflora, and P. obovate. P. lactiflora ‘Lv He’ is sister to the clade of P. lactiflora.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Acemel RD, Tena JJ, Irastorza-Azcarate I, Marlétaz F, Gómez-Marín C, de la Calle-Mustienes E, Bertrand S, Diaz SG, Aldea D, Aury J-M, et al. 2016. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate hox bimodal regulation. Nat Genet. 48(3):336–341.
- Eason J, Pinkney T, Heyes J, Brash D, Bycroft B. 2002. Effect of storage temperature and harvest bud maturity on bud opening and vase life of Paeonia lactiflora cultivars. NZJ Crop Hortic Sci. 30(1):61–67.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Guo S, Guo LL, Zhao W, Xu J, Li YY, Zhang XY, Shen XF, Wu ML, Hou XG. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules. 23(2):246.
- Li L, Cheng FY, Zhang QX. 2011. Microsatellite markers for the Chinese herbaceous peony Paeonia lactiflora (Paeoniaceae). Am J Bot. 98(2):e16–e18.
- Lowe T M, Eddy S R. 1997. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Research. 25(5):955–964. doi:10.1093/nar/25.5.955.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):2047–217X-1-18.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.
- Yu XL, Tan W, Zhang HY, Gao H, Wang WX, Tian XX. 2019. Complete chloroplast genomes of Ampelopsis humulifolia and Ampelopsis japonica: Molecular structure, comparative analysis, and phylogenetic analysis. Plants. 8(10):410.
