Abstract
The complete chloroplast genome sequence of a wild Paeonia lactiflora species from Henan province, Paeonia obovata subsp. Willmottiae (Stapf) D. Y. Hong et K. Y. Pan, was applied to support the difference of Paeonia lactiflora by next-generation sequencing. The length of complete chloroplast genome is 152,660 bp, including a large single-copy (LSC) region, a small single-copy (SSC) region, and a pair of identical inverted repeat (IR) regions (IRa and IRb) with length of 84,363 bp, 16,983 bp and 25,657 bp, respectively. Altogether, 115 genes in chloroplast genome were annotated including 81 protein-encoding genes, 30 tRNA genes, and 4 rRNA genes. Paeonia obovata subsp. Willmottiae (Stapf) D. Y. Hong et K. Y. Pan clustered in the clade of Paeonia lactiflora showed a closer relationship with Paeonia sect. Mudan rather than Paeonia Brownii.
Paeonia lactiflora is an important herbaceous species of which the seeds contain edible oil and total monoterpene glycosides (Liu et al. Citation2017). The chloroplast genome of paeonia lactiflora has become a researching hotspot in recent years (Samigullin et al. Citation2018; Lee et al. Citation2019; Gao et al. Citation2020; Zhang et al. Citation2020). Paeonia obovata subsp. Willmottiae (Stapf) D. Y. Hong et K. Y. Pan is a wild Paeonia lactiflora species distributed in the northeastern part of Sichuan, southern Gansu, southern Shaanxi, western Hubei, Henan (Song County) and Anhui (Jiuhua Mountain). It usually grows under forests on hillside at an altitude of 1500–2000 m (Hong et al. Citation2001). Study of complete chloroplast genome of this wild species from Henan province might provide useful biological information for maintaining the distinction among the species in the genus Paeonia.
Leaves were collected and deposited at the National Peony Gene Bank of Luoyang, Henan, China (N34°42′29.70″, E112°23′36.14″). The specimen of Paeonia obovata subsp. Willmottiae (Stapf) D. Y. Hong et K. Y. Pan is stored in the Paeonia Specimen Museum of Henan University of Science and Technology under accession number PL2018041502. Total genomic DNA was extracted using the CTAB method (Guo et al. Citation2018). Genomic DNA purity was tested by Qubit 2.0 and NanoDrop. DNA quality was accessed by agarose gel electrophoresis. Illumina X ten platform was used to construct sequencing libraries. Chloroplast genome was assembled using the mainstream software including FastQC (Version 0.11.5), Skewer (Version 0.2.2) (Jiang et al. Citation2014), seqkit (Version v0.9.0) (Shen et al. Citation2016) and ABYSS (Version 2.0.0) (Simpson et al. Citation2009). CPGAVAS platform was used for the annotation of the chloroplast genome. The OGDRAW platform was used for the gene map completion. The chloroplast genome sequences were submitted to GenBank under the accession number of MN149613.
The length of complete chloroplast genome of Paeonia obovata subsp. Willmottiae (Stapf) D. Y. Hong et K. Y. Pan is 152,660 bp and GC content is 38.4%. The complete chloroplast genome contains four regions: a large single-copy (LSC) region, a small single-copy (SSC) region, and a pair of identical inverted repeat (IR) regions. The length of LSC is 84,363 bp and its GC content is 36.7%. The length of SSC is 16,983 bp and its GC content is 32.8%. The length of IR is 25,657 bp and its GC content is 43.1%. A total of 115 genes were annotated, of which 81 protein-encoding genes, 30 tRNA genes, and 4 rRNA genes were detected.
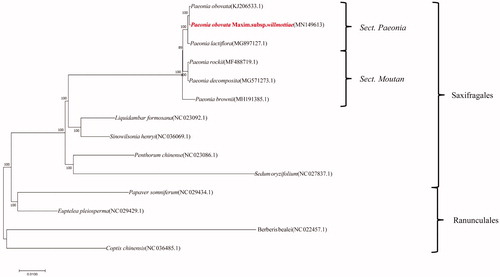
The complete chloroplast genome sequences of 14 species from Saxifragales and Ranunculales were used to understand the phylogenetic relationship and genetic diversity (). The neighbour-joining phylogenetic trees constructed by MAGA software (Version 5.2) (Tamura et al. Citation2011) manifests that Paeonia obovata subsp. Willmottiae (Stapf) D. Y. Hong et K. Y. Pan appeared as a sister to the clade of Paeonia lactiflora. Paeonia obovata was clustered into the same clade with Paeonia sect. Mudan, showing a closer relationship with Paeonia sect. Mudan rather than Paeonia Brownii.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Gao CS, Wang QR, Ying ZQ, Ge YQ, Cheng RB. 2020. Molecular structure and phylogenetic analysis of complete chloroplast genomes of medicinal species Paeonia lactiflora from Zhejiang province. Mitochondrial DNA Part B. 5(1):1077–1078.
- Guo S, Guo LL, Zhao W, Xu J, Li YY, Zhang XY, Shen XF, Wu ML, Hou XG. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules. 23(2):246.
- Hong DY, Pan KY, Rao GY. 2001. Cytogeography and taxonomy of the Paeonia obovata polyploid complex (Paeoniaceae). Plant Syst Evol. 227(3–4):123–136.
- Jiang HS, Rong L, Ding SW, Zhu SF. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf. 15(1):182.
- Lee M, Park JH, Gil J, Lee JH, Lee Y. 2019. The complete chloroplast genome of Paeonia lactiflora Pall. (Paeoniaceae). Mitochondrial DNA Part B. 4(2):2715–2716.
- Liu P, Xu YF, Gao XD, Zhu XY, Du MZ, Wang YX, Deng RX, Gao JY. 2017. Optimization of ultrasonic-assisted extraction of oil from the seed kernels and isolation of monoterpene glycosides from the oil residue of Paeonia lactiflora Pall. Ind Crops Prod. 107:260–270.
- Samigullin T H, Logacheva M D, Degtjareva G V, Efimov S V, Terentieva E I, Vallejo-Roman C M. 2018. Complete plastome sequence of Paeonia lactiflora Pall. (Paeoniaceae: Saxifragales). Mitochondrial DNA Part B. 3(2):1110–1111. doi:10.1080/23802359.2018.1501311.
- Shen W, Le S, Li Y, Hu FQ. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLOS One. 11(10):e0163962.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.
- Zhang MY, Li YM, Gao J, Chen Y, Yan YG, Zhang YQ, Luo Y, Zhang G. 2020. Complete plastid genome of the Chinese medicinal herb Paeonia obovata subsp. Willmottiae (Paeoniaceae): characterization and phylogeny. Mitochondrial DNA Part B. 5(1):845–847.