Abstract
Gardenia jasminoides is well distributed and planted in China because of high ornamental and medical applications in Rubiaceae. Therefore, aiming at studying the biodiversity and solar-related genes, we acquired its complete chloroplast genome. The chloroplast genome was 155,096 bp in length with typical circle structure, it presented an 85,409 bp large single-copy (LSC) region, an 18,103 bp small single-copy (SSC) region, and two 25,792 bp inverted repeat (IRs) regions. The overall GC content was 37.49%, SSC and IR components showed extreme GC percentages ranging from 31.53 to 43.18. A brief online annotation reported 125 CDSs, 8 rRNAs, and 33 tRNA. To our surprise, G. jasminoides was closer to Coffea canephora and Coffea arabica within Rubiaceae from phylogenetic analysis. This chloroplast genome announcement of the G. jasminoides could embody light on the artificial breeding and further researches in the genus Gardenia Ellis and recruit more studies in this family.
Gardenia jasminoides, also known as Cape Jasmine, is an evergreen shrub found in tropical and semitropical countries. Also, white flowers with strong fragrance make it enjoyable all round the world. It is positioned at Subfam. Cinchonoideae of Rubiaceae in the traditional taxonomy. In south China, G. jasminoides blooms in warm-climate months from late spring to summer, fruits with wing-like longitudinal ribs make it recognizable out of five indigenous species, besides thick, lustrous, lance-shaped leaves. This species can be easily found in south Chinese provinces, especially in Jiangxi, Hunan, and Fujian, there are abundant wild resources and artificial cultivation in these areas. Gardenia Jasminoides is well planted for three main purpose. Its gorgeous flowers make living environments welcome. The dried fruits are both used as traditional medicine and natural food pigment.
Rubiaceae owns over 10 thousand species as reported, many species demonstrate important economic uses. With the dramatic promotion of sequencing technology, researchers are trying to solve the mysteries lying deep in the plant genomes. To date, the coffee genome has been a pioneer in the exploration of genome-level biodiversity and biotin biosynthesis in Rubiaceae (Denoeud et al. Citation2014; Guyeux et al. Citation2019). Besides, the chloroplast genomes about 19 species in different genus of Rubiaceae have been reported to answer the puzzles among nearby families as one of the three genetic systems of plants. Thus, focusing on the dual characteristics of sequence variation and conservation, we reported a high-quality chloroplast genome of G. jasminoides to study chloroplast genomics. We also generated comparisons of related species to make the evolution of Rubiaceae clear by chloroplast super barcodes.
Wide-type plants of G. jasminoides were originally collected by Dr. Zhou from City Quanzhou, Fujian province, China, and experimental samples were then kept in the laboratory of Fujian Agriculture and Forestry University. Total genomic DNA was extracted by the modified CTAB method to reach the quantity and quality for further sequencing. The frozen samples including fresh tissues, specimens and sequenced DNA were stored in the laboratory of Fujian Agriculture and Forestry University (Voucher specimen: ZZ-FJ2019-13A, FAFU, 23°32′25.19″N 120°47′57.71″E). 6 Gb pair-end clean reads were obtained by PE150 library strategy and the BGI-500 platform (BGI, Wuhan, China) (Mak et al. Citation2017). Adapters and low-quality reads were removed automatically by fastp software (Chen et al. Citation2018). Then, the processed data were assembled by GetOrganelle v1.5.2 flow, in which core mapping software and assembly tool used were bowtie2 and Spades version 3.13.1 (Vasilinetc et al. Citation2015). In this flow, random separated reads were rectified and assembled into contigs. In the following steps, fragments with low-sequence coverages were removed as noises during the screening progress by using Bandagev0.8.1 (Wick et al. Citation2015) and ultimately formed a high-coverage circle chloroplast. Then, the initial clean reads were mapped to the draft genome to inspect the assembling consistency. Then, this genome was preliminarily annotated with large single copy region using Geneious Prime to adjust the starting position. The nucleotide information in detail was counted by Bioedit. As a result, we established a length of 155,096 bp circle chloroplast genome of G. jasminoides with a total GC content of 37.49%. This cp genome typical includes a length of 85,409 bp large single-copy (LSC) region and an 18,103 bp small single-copy (SSC) region, separated by two 25 792 bp inverted repeat (IRs) regions. LSC and SSC, 35.31% and 31.53%, are interrupted by two 43.18% GC content IRs from both sides. After assessment of the assembled plastid genome, annotation of the new plastid genome was conducted by online software GeSeq and third-party annotators like ARAGORN v1.2.38, tRNAscan-SE v2.0.5 against embryophyta chloroplast database. In this chloroplast genome, 33 tRNA and 8 conserved rRNA were found, respectively. 125 CDSs were annotated by HMMER. The assembled cp genome sequence of G. jasminoides can be detected in GenBank with an accession number of MT018450.
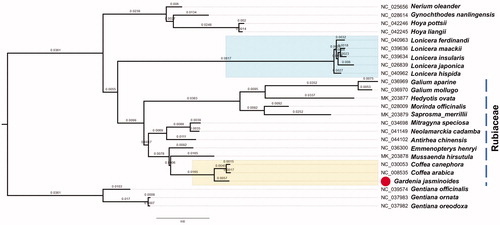
Though there exit part sequenced species, more genetic information about this family is needed (Bruy et al. Citation2018; Ehrendorfer et al. Citation2018). We collected most species’ chloroplast genome in Rubiaceae, trying our best to get a clear phylogenetic relationship. Thus, we rebuilt a phylogenetic tree with the same method as before. Alignment was done following HomBlocks pipeline (Bi et al. Citation2018). We reconstructed a phylogeny employing the GTRCAT model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference by means of RAxML-HPC v.8.2.10 on the CIPRES cluster. The new ML tree () was consistent with recent phylogenetic results in Rubiaceae(Ehrendorfer et al. Citation2018), 3 species in Gentiana genus were chosen as outgroup taxa. However, these relationships of Rubiaceae still reflected problems that current taxonomy does not fit the phylogenetic relationships, and is disturbed by nearby families. Gardenia jasminoides displayed a closer kinship to coffee, which both include Coffea canephora and Coffea Arabica within the family Rubiaceae. Most species used in Rubiaceae fell into one main branche, and Lonicera displayed a genetic intimacy in terms of chloroplast super barcodes. Here, we believe this presentation of G. jasminoides chloroplast genome helps clarify its evolutionary status in genus Gardenia, especially among nearby cultivars, and provides genomic resources for artificial breeding and fundamental researches.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of selected chloroplast sequences. The main branch that contain 13 species in Rubiaceae is marked with dotted line. Gardenia jasminoides (MT018450) is marked with red circle. Genebank accession numbers were listed before their corresponding species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Bruy D, Hattermann T, Barrabe L, Mouly A, Barthelemy D, Isnard S. 2018. Evolution of plant architecture, functional diversification and divergent evolution in the genus Atractocarpus (Rubiaceae) for New Caledonia. Front Plant Sci. 9(1775). doi:10.3389/fpls.2018.01775
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 345(6201):1181–1184.
- Ehrendorfer F, Barfuss MHJ, Manen JF, Schneeweiss GM. 2018. Phylogeny, character evolution and spatiotemporal diversification of the species-rich and world-wide distributed tribe Rubieae (Rubiaceae). PLoS One. 13(12):e0207615.
- Guyeux C, Charr JC, Tran HTM, Furtado A, Henry RJ, Crouzillat D, Guyot R, Hamon P. 2019. Evaluation of chloroplast genome annotation tools and application to analysis of the evolution of coffee species. PLoS One. 14(6):e0216347.
- Mak SST, Gopalakrishnan S, Caroe C, Geng C, Liu S, Sinding MS, Kuderna LFK, Zhang W, Fu S, Vieira FG, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 6(8):1–13.
- Vasilinetc I, Prjibelski AD, Gurevich A, Korobeynikov A, Pevzner PA. 2015. Assembling short reads from jumping libraries with large insert sizes. Bioinformatics. 31(20):3262–3268.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
