Abstract
We report the complete mitochondrial genome of Eudarcia gwangneungensis Roh & Byun, Citation2019. The genome has a total length of 15,391 bp (GenBank accession number: MN413148), consisting of 13 protein-coding genes, 22 tRNA, 2 rRNA genes, and A + T-rich control region. The nucleotide composition was 39.4% T, 40.4% A, 12.7% C, and 7.4% G. This study is the first report of a complete mitochondrial genome of a meessiid species, and the mitogenomic sequence can be used as a reference data for extensive phylogenetic studies of Tineoidea.
Family Meessiidae has been previously placed as a subfamily in Tineidae. Regier et al. (Citation2013) treated most species of Tineidae by dividing into two groups: namely, most species in Tineidae 1, and members of Eudarcia Clemens, Citation1860 as Tineidae 2. Recently, Regier et al. (Citation2015) revised the classification system of Tineoidea and revived two families, namely Dryadaulidae and Meessiidae, based on a molecular phylogenetic analysis. Family Meessiidae consists of two genera, Eudarcia Clemens, Citation1860 and Bathroxena Meyrick, Citation1919 (Regier et al. Citation2015). The genus Eudarcia was established with the type species Eudarcia simulatricella Clemens, Citation1860 and currently includes 77 species (Bidzilya et al. Citation2016; Budashkin and Bidzilya Citation2018). Recently, Roh and Byun (Citation2019) report the family Meessiidae for the first time from Korea: three new species of the genus Eudarcia: E. gwangneungensis Roh and Byun, Citation2019, E. longiphalla Roh and Byun, Citation2019, and E. cornea Roh and Byun, Citation2019, plus one unrecorded species, E. orbiculidomus (Sakai and Saigusa Citation1999).
The specimens of E. gwangneungensis were collected from Gwangneung forest, Pocheon, South Korea (37.752619, 127.585850) in March 2019, which are deposited in the Systematic Entomology Laboratory, Hannam University (SEL/HNU), Daejeon, South Korea (sample number: HNUE-RSJ161). DNA was sequenced with Illumina Hiseq (Macrogen, Inc., Seoul, Korea). A total of 13,269,560 reads were analyzed to generate 2,003,703,560 base pairs of sequence and assembled in Geneious 11.0.5 (Kearse et al. Citation2012). The gene annotation was accomplished and circularity was checked with MITOS2 webserver (Bernt et al. Citation2013, http://mitos.bioinf.uni-leipzig.de/).
The complete mitochondrial genome sequence of E. gwangneungensis (GenBank accession number: MN413148) has a total length of 15,391 bp, consisting of 13 protein-coding genes, 22 tRNA, 2 rRNA genes, and an AT-rich control region. The mitogenome contains 39.4% T, 40.4% A, 12.7% C, and 7.4% G, which showed a high A + T. All of the PCGs started with ATN as the start codon. The A + T-rich region is of the full length 416 bp.
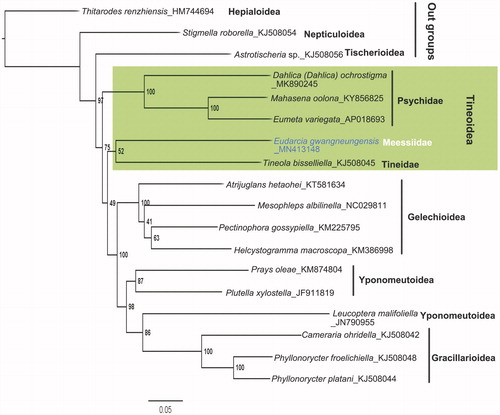
In this study, the phylogenetic relationship of E. gwangneungensis was estimated using maximum likelihood analysis in RAxML8.1.2 (Stamatakis Citation2014). Also, the ML tree was constructed with the amino acid sequences of the 13 PCGs from 18 species in eight superfamilies, including five species from Tineoidea (used as ingroups) and 10 species from three superfamilies (used as sister groups). Among them, three species from Hepialoidea, Nepticuloidea, and Tischerioidea were selected as outgroups. The result of ML analysis () shows that E. gwangneungensis are distinctly included in the family Meesiidae within Tineoidea positioned as a sister taxon of Tineoidea. It may reflect the recent classification of Tineoidea (Regier et al. Citation2015), even if the support value of the clade formed by meesiid and tineid species represents a lower result.
Figure 1. Phylogenetic tree based on the maximum-likelihood analysis of concatenated amino acid sequences of 13 mitochondrial protein-coding genes of E. gwangneungensis and other glossatan species using Geneious 11.0.5. The numbers beside the nodes indicate percentages of 1000 bootstrap values. Thitarodes renzhiensis (Hepialoidea), Stigmella roborella (Nepticuloidea), and Astrotischeria sp. (Tischerioidea) were used as outgroups. Alphanumeric terms indicate the GenBank accession numbers.
Nucleotide sequence accession number: The complete mitochondrial genome sequence of Eudarcia gwangneungensis has been assigned GenBank accession number MN413148.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bidzilya OV, Budashkin YI, Gaedike R. 2016. A revision of Eudarcia glaseri-species group (Lepidoptera, Meessiidae) with description of two new species from Greece and Crimea. Zootaxa. 4179(3):547–560.
- Budashkin Y, Bidzilya O. 2018. Four new species of the genus Eudarcia Clemens, 1880 (Lepidoptera: Meessiidae) from Crimea. Zootaxa. 4446(1):111–124.
- Clemens JB. 1860. Contributions to American lepidopterology. Proceedings of the Academy of Natural Sciences; Philadelphia; Vol. 12; p. 4–15, 156. 174, 203–221, 345–362, 522–547.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Meyrick E. 1919. Exotic Microlepidoptera. Vol. 2. London (UK): Taylor and Francis.
- Regier J, Mitter C, Davis DR, Harrison T, Sohn JC, Cummings M, Zwick A, Mitter K. 2015. A molecular phylogeny and revised classification for the oldest ditrysian moth lineages (Lepidoptera: Tineoidea): were the megadiverse Ditrysia ancestrally non-phytophagous? Syst Entomol. 40(2):409–432.
- Regier JC, Mitter C, Zwick A, Bazinet AL, Cummings MP, Kawahara AY, Sohn JC, Zwickl DJ, Cho S, Davis DR, et al. 2013. A large-scale, higher-level, molecular phylogenetic study of the insect order Lepidoptera (moths and butterflies). PLoS One. 8(3):e58568.
- Roh SJ, Byun BK. 2019. The Meessiidae (Lepidoptera: Tineoidea) of Korea. Florida Entomol. 102:65–75.
- Sakai M, Saigusa T. 1999. A new species of Obesoceras Petersen, 1957 from Japan (Lepidoptera: Tineidae). Entomol Sci. 2:405–412.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
