Abstract
Salvia hispanica is an annual food and oil crop native to Mexico and Guatemala. In this study, we report the complete chloroplast genome (plastome) of S. hispanica. The plastome was 150,980 bp in length and comprises a large single-copy region (82,274 bp), a small single-copy region (17,534 bp), and a pair of inverted repeats (25,586 bp). It encodes 114 unique genes, including 80 protein-coding genes (PCGs), 30 tRNAs, and 4 rRNAs. The overall GC content of this plastome was 38.0%. Phylogenomic analysis of all PCGs revealed that S. hispanica was closely related to S. bulleyana.
Keywords:
Salvia, with nearly 1000 species, belongs to Lamiaceae. It is radiated extensively in three regions of the world: Central and South America, Western Asia, and Eastern Asia (Walker and Sytsma Citation2007). Salvia hispanica is an annual herb native to Mexico and Guatemala (Cahill and Provance Citation2002). It has a long history of edible cultivation and is the third largest food crop in Mexico and other places after corn and soybeans (Muñoz et al. Citation2013). Its seeds are rich in unsaturated fatty acid, high-quality protein, soluble dietary fiber, vitamins, mineral elements, and a variety of antioxidant compounds. It has significant preventive and therapeutic effects on obesity, cardiovascular disease, and diabetes (Reyes-Caudillo et al. Citation2008; Alfredo et al. Citation2009; Ixtaina et al. Citation2011; Poudyal et al. Citation2012). They also have anti-inflammatory and anti-atherosclerotic effects (da Silva et al. Citation2019; Grancieri et al. Citation2019). Its serine-type protease inhibitors have shown the most positive effects with hepatocarcinoma (HCC) model (Laparra and Haros Citation2019). In this study, we reported the plastome of S. hispanica for resolving its phylogenetic position.
Fresh leaves of S. hispanica were collected from Kunming Botanical Garden, Chinese Academy of Sciences (N25°08′33.70″, E102°44′30.73″). Voucher specimen (SD118) has been deposited at College of Life Sciences, Shandong Normal University. Total genomic DNA was extracted by the modified CTAB method described in Wang et al. (Citation2013). The total genomic DNA was used for library preparation and paired-end (PE) sequencing by the Illumina MiSeq instrument at Novogene (Beijing, China). The plastome was assembled using Organelle Genome Assembler (OGA, https://github.com/quxiaojian/OGA) described in Qu (Citation2019). Plastome annotation was conducted with Plastid Genome Annotator (PGA, https://github.com/quxiaojian/PGA) (Qu et al. Citation2019), coupled with manual correction using Geneious v9.1.4. To determine the phylogenetic placement of S. hispanica, a maximum-likelihood (ML) tree was reconstructed using RAxML v8.2.10 (Stamatakis Citation2014), including tree robustness assessment using 1000 rapid bootstrap replicates with the GTRGAMMA substitution model, based on the alignment of 80 shared protein-coding genes (PCGs) using MAFFT v7.313 (Katoh and Standley Citation2013).
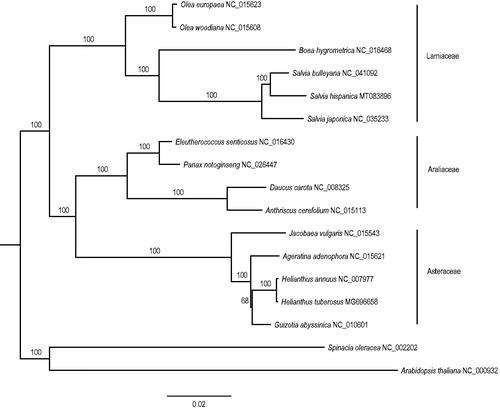
The complete plastome of S. hispanica (GenBank accession number: MT083896) was 150,980 bp in size and contained a large single-copy region (LSC: 82,274 bp), a small single-copy region (SSC: 17,534 bp), and two inverted repeats regions (IR: 25,586 bp). The overall GC content was 38.0%. In total, 114 unique genes, including 80 PCGs, 30 tRNAs, and 4 rRNAs were annotated. Among them, 14 PCGs and 6 tRNAs contained introns, in which 12 PCGs and 6 tRNAs contained one intron and 2 PCGs contained two introns. There were 11 duplicated genes in the IR. The ML phylogenetic tree showed that S. hispanica was closely related to S. bulleyana ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alfredo VO, Gabriel RR, Luis CG, David BA. 2009. Physicochemical properties of a fibrous fraction from chia (Salvia hispanica L.). LWT – -Food Sci Technol. 42(1):168–173.
- Cahill JP, Provance MC. 2002. Genetics of qualitative traits in domesticated chia (Salvia hispanica L.). Heredity. 93(1):52–55.
- da Silva BP, Toledo RCL, Mishima MDV, Moreira MEdC, Vasconcelos CM, Pereira CER, Favarato LSC, Costa NMB, Martino HSD. 2019. Effects of chia (Salvia hispanica L.) on oxidative stress and inflammation in ovariectomized adult female wistar rats. Food Funct. 10(7):4036–4045.
- Grancieri M, Martino HSD, Mejia E. 2019. Chia (Salvia hispanica L.) seed total protein and protein fractions digests reduce biomarkers of inflammation and atherosclerosis in macrophages in vitro. Mol Nutr Food Res. 63(19):1900021.
- Ixtaina VY, Martínez ML, Spotorno V, Mateo CM, Maestri DM, Diehl BWK, Nolasco SM, Tomás MC. 2011. Characterization of chia seed oils obtained by pressing and solvent extraction. Food Compos Anal. 24(2):166–174.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Laparra JM, Haros CM. 2019. Plant seed protease inhibitors differentially affect innate immunity in a tumor microenvironment to control hepatocarcinoma. Food Funct. 10(7):4210–4219.
- Muñoz LA, Cobos A, Diaz O, Aguilera JM. 2013. Chia seed (Salvia hispanica): an ancient grain and a new functional food. Food Rev Int. 29(4):394–408.
- Poudyal H, Panchal SK, Waanders J, Ward L, Brown L. 2012. Lipid redistribution by α-linolenic acid-rich chia seed inhibits stearoyl-CoA desaturase-1 and induces cardiac and hepatic protection in diet-induced obese rats. Nutr Biochem. 23(2):153–162.
- Qu XJ. 2019. Complete plastome sequence of an endangered species, Calocedrus rupestris (Cupressaceae). Mitochondrial DNA B. 4(1):762–763.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Reyes-Caudillo E, Tecante A, Valdivia-López MA. 2008. Dietary fibre content and antioxidant activity of phenolic compounds present in Mexican chia (Salvia hispanica L.) seeds. Food Chem. 107(2):656–663.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Walker JB, Sytsma KJ. 2007. Staminal evolution in the genus Salvia (Lamiaceae): molecular phylogenetic evidence for multiple origins of the staminal lever. Ann Bot. 100(2):375–391.
- Wang HY, Jiang DF, Huang YH, Wang PM, Li T. 2013. Study on the phylogeny of Nephroma helveticum and allied species. Mycotaxon. 125(1):263–275.