Abstract
Pyropia dentata is an economically valuable species in seaweed aquaculture in the southwest coastal regions of Korea. Herein, we determined the complete chloroplast (cp) genome sequence of P. dentata using PacBio resequencing data. The whole cp genome is 192,266 bp in size, encodes 148 genes (including 92 protein-coding, 44 tRNA-coding, and 12 rRNA-coding genes), and lacks inverted repeat (IR) regions. A maximum likelihood phylogenetic analysis demonstrated that P. dentata was most closely related to P. haitanensis. The cp genome will provide a useful reference for further studies of P. dentata.
Pyropia, a genus of red algae in the Bangiaceae family, is one of the most economically important marine crops in East Asian countries such as South Korea, China, and Japan. The production of Pyropia as a marine crop is valued at over $2 billion per year (Blouin et al. Citation2011). Pyropia dentata is a native species that is distributed and intensively farmed in the southwest coastal regions of Korea. Although P. dentata production represents 10–20% of the total production of cultivated Pyropia in Korea, this species is increasing in economic value in Korea due to its plasticity in response to climate change (Kim et al. Citation2017). To gain insight into its evolution and facilitate genetic research into this important Pyropia species, we characterized the complete chloroplast (cp) genome of P. dentata based on PacBio sequencing data.
Young blade samples were collected from Songji-myeon (36°23′35.54″N, 26°28′23.45″E), Haenam County, Jeonnam Province, Republic of Korea, and deposited in the Ocean and Fisheries Science Institute Haenam Branch at Haenam, Korea (specimen code JOFP0004562). Genomic DNA was extracted from young blades using a DNeasy Power Plant Pro Kit (Qiagen, Valencia, CA), in accordance with the manufacturer’s protocol. Whole-genome resequencing was performed at Coscience Co. Ltd. (Mokpo, South Korea) using two single-molecule real time (SMRT) cells on a PacBio RS II sequencing platform (Pacific Biosciences, Menlo Park, CA). The raw data were filtered using the RS Filter Only protocol in the SMRT portal (Pacific Biosciences, Menlo Park, CA) with default settings. Short contigs of <500 nucleotides in length were discarded. Sequences were assembled using a Celera Assembler v8.3 (Myers et al. Citation2000) with the cp genome of the closely related species Porphyra pulchra (NC_029861) as the reference. Finally, the assembled cp genome was annotated and adjusted manually using DOGMA (Wyman et al. Citation2004). The annotated cp genome of P. dentata was submitted to GenBank under the accession number LC521919.
The complete cp genome of P. dentata is a circular molecule that is 192,266 bp in length and has an overall GC content of 32.6%. However, no typical inverted repeat (IR) regions are present, which would make it challenging to define large single-copy (LSC) and small single-copy (SSC) regions. The new sequence has a total of 148 genes, including 92 protein-coding genes, 44 tRNA-coding genes, and 12 rRNA-coding genes.
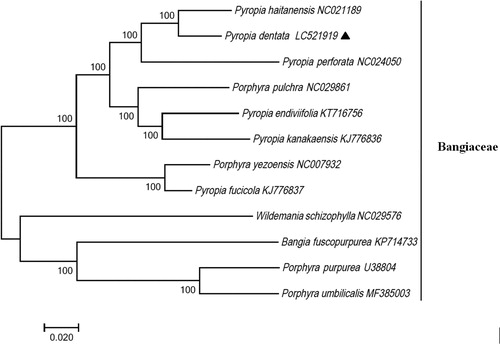
To investigate its phylogenetic position, the sequences of 11 complete cp genomes of Bangiaceae (P. umbilicalis, P. pulchra, P. endiviifolia, P. purpurea, P. fucicola, P. perforata, P. yezoensis, P. haitanensis, P. kanakaensis, Bangia fuscopurpurea, and Wildemania schizophylla) were aligned using Mega 7.0 (Kumar et al. Citation2016) with 1000 bootstrap replicates. All the data were downloaded from NCBI GenBank. Phylogenetic analysis based on the complete cp genomes showed that P. dentata was most closely related to P. haitanensis (). We believe that the cp genome of P. dentata is a valuable tool for studies of species identification, phylogenetics, and genetic diversity of Bangiaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Blouin NA, Brodie JA, Grossman AC, Xu P, Brawley SH. 2011. Porphyra: a marine crop shaped by stress. Trends Plant Sci. 16(1):29–37.
- Kim JK, Yarish C, Hwang EK, Park M, Kim Y. 2017. Seaweed aquaculture: cultivation technologies, challenges and its ecosystem services. Algae. 32(1):1–13.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. 2000. A whole-genome assembly of Drosophila. Science. 287(5461):2196–2204.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.