Abstract
We describe the plastome of Jenufa minuta, a unicellular green alga whose precise relationship with the Sphaeropleales (Chlorophyceae) remains uncertain. This 206,680-bp genome contains 100 conserved genes, 11 of which exhibit 16 group I and seven group II introns. We discovered a trans-spliced group II intron in the rpl32 gene of J. minuta and its orthologs in all Sphaeropleales and several Chlamydomonadales; this is the first time this intron is documented. As in the mitochondria of J. minuta, the UGA codon is decoded as tryptophan. To our knowledge, this is the second report of a variant genetic code in plastids.
Departures from the standard genetic code arose more frequently in mitochondria than in plastids during the evolution of photosynthetic algae. Among green algae, various noncanonical genetic codes have been observed in mitochondria, particularly in compact mitogenomes that underwent reduction in gene content; e.g. in those of the prasinophyte Pycnococcus provasolii (Turmel et al. Citation2010), representatives of the Pedinomonadales and Marsupiomonadales (Pedinophyceae) (Turmel et al. Citation1999; Turmel, Otis, Lemieux Citation2020), and several members of the Sphaeropleales (Chlorophyceae) (Noutahi et al. Citation2019; Zihala and Elias Citation2019). In contrast, evidence for a variant genetic code (i.e. reassignment of AUA from isoleucine to methionine) in the plastome has been reported only for the tiny prasinophytes belonging to the genus Chloroparvula (Chloropicophyceae) (Turmel et al. Citation2019). Here, we present the plastome sequence of Jenufa minuta (Chlorophyceae, incertae sedis) and provide evidence for a second case of noncanonical genetic code. This chlorophycean alga, whose mitogenome also exhibits a variant genetic code (Turmel, Otis, Vincent, et al. Citation2020), belongs to a lineage that is sister to Sphaeropleales or represents a deep lineage of the latter order (Lemieux et al. Citation2015).
J. minuta (CAUP H8102) was obtained from the Culture Collection of Algae at Charles University in Prague. Total cellular DNA was sequenced on the Roche 454 platform by the Genomic Analysis Platform of Laval University. The methods of DNA extraction, sequencing and assembly of the plastome sequence have been described previously (Lemieux et al. Citation2015).
The 206,680-bp plastome of J. minuta (GenBank NC_028582) displays a 11,100-bp inverted repeat that contains only the rDNA operon. The repertoire of 100 different conserved genes comprises 29 tRNA and 68 protein-coding genes (including rpl12, a gene present in Sphaeropleales but missing in other chlorophycean orders). Coding regions of 11 genes are interrupted by 16 cis-spliced group I introns and seven group II introns, three of which are trans-spliced. Two of the latter introns occur in psaA and the other in rpl32 only 13 nucleotides downstream of the initiator codon. This is the first time that a trans-spliced intron is reported in rpl32 among green algae. Upon reanalysis of complete chlorophycean plastomes, we identified this trans-spliced intron in all Sphaeropleales and several Chlamydomonadales. Modeling of the J. minuta intron secondary structure revealed that the breakpoint falls within domain IV.
The UGA codon is a sense codon in the J. minuta plastid. It resides at 64 positions within the coding regions of 12 protein-coding genes and is decoded as tryptophan according to the alignment of the protein products against their green algal homologs. UGA is also decoded as tryptophan in the mitochondria of J. minuta and Jenufa perforata (Turmel, Otis, Vincent, et al. Citation2020). The trnW(uca) gene was not detected in the J. minuta plastome; hence, it is unclear which tRNA recognizes the UGA codon in the plastid.
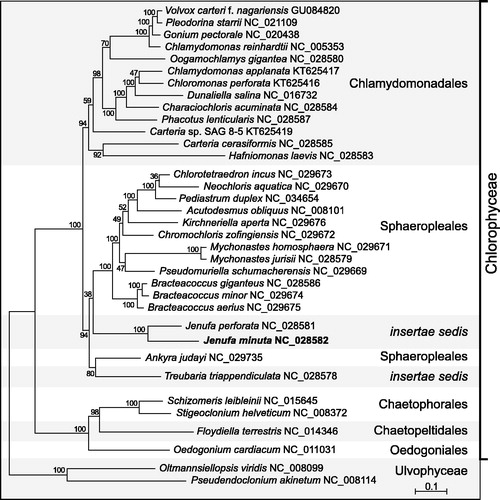
Phylogenetic analysis of 55 plastome-encoded proteins from 33 chlorophycean algae using RAxML v.8.2.3 (Stamatakis Citation2014) recovered a strongly supported clade containing J. minuta and J. perforata, which is sister to most Sphaeropleales ().
Figure 1. RAxML tree inferred from 55 concatenated plastome-encoded proteins of 33 chlorophycean green algae. Oltmannsiellopsis viridis and Pseudendoclonium akinetum (Ulvophyceae) were used as outgroup taxa. The figure shows the best-scoring tree, with bootstrap support values (100 replicates) reported on the nodes. GenBank accession numbers are provided for the plastomes of all taxa. The scale bar denotes the estimated number of amino acid substitutions per site. The data set was generated using the predicted protein sequences derived from atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, clpP, petB, petD, petG, petL, psaA, psaB, psaC, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, rbcL, rpl2, rpl5, rpl14, rpl16, rpl20, rpl23, rpl36, rpoA, rpoB, rpoC2, rps3, rps4, rps7, rps8, rps9, rps11, rps12, rps14, rps18, rps19, tufA, ycf3. Following alignment of the sequences of individual proteins with Muscle v3.7 (Edgar Citation2004), ambiguously aligned regions were removed using TrimAL v1.4 (Capella-Gutierrez et al. Citation2009) with the options block = 6, gt = 0.7, st = 0.005 and sw = 3, and the protein alignments were concatenated using Phyutility v2.2.6 (Smith and Dunn Citation2008). The phylogenetic analysis was carried out under the GTR + Γ4 model.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M. 2015. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol. 15(1):264.
- Noutahi E, Calderon V, Blanchette M, El-Mabrouk N, Lang BF. 2019. Rapid genetic code evolution in green algal mitochondrial genomes. Mol Biol E. 36(4):766–783.
- Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Turmel M, Lemieux C, Burger G, Lang BF, Otis C, Plante I, Gray MW. 1999. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor: two radically different evolutionary patterns within green algae. Plant Cell. 11(9):1717–1730.
- Turmel M, Lopes Dos Santos A, Otis C, Sergerie R, Lemieux C. 2019. Tracing the evolution of the plastome and mitogenome in the Chloropicophyceae uncovered convergent tRNA gene losses and a variant plastid genetic code. Genome Biol E. 11(4):1275–1292.
- Turmel M, Otis C, Lemieux C. 2010. A deviant genetic code in the reduced mitochondrial genome of the picoplanktonic green alga Pycnococcus provasolii. J Mol Evol. 70(2):203–214.
- Turmel M, Otis C, Lemieux C. 2020. Complete mitogenome of the chlorophyte green alga Marsupiomonas sp. NIES 1824 (Pedinophyceae). Mitochondrial DNA Part B. 5(1):548–550.
- Turmel M, Otis C, Vincent AT, Lemieux C. 2020. The complete mitogenomes of the green algae Jenufa minuta and Jenufa perforata (Chlorophyceae, incertae sedis) reveal a variant genetic code previously unrecognized in the Chlorophyceae. Mitochondrial DNA Part B. 5(2):1516–1518.
- Zihala D, Elias M. 2019. Evolution and unprecedented variants of the mitochondrial genetic code in a lineage of green algae. Genome Biol E. 11(10):2992–3007.
