Abstract
Vitis vinifera ‘Kyoho’ is one kind of table grape cultivar with exquisite flavor, color, and health benefits. The length of complete chloroplast genome sequence of Vitis vinifera ‘Kyoho’ is 160,928 bp and its GC content is 37.4%. The length of LSC is 89,148 bp and its GC content is 35.3%. The length of SSC is 19,072 bp and its GC content is 31.7%. The length of IR is 26,354 bp and its GC content is 43.0%. There were 113 genes annotated including 80 protein-encoding genes, 29 tRNA genes, and 4 rRNA genes. The neighbor-joining phylogenetic trees indicated that Vitis vinifera ‘Kyoho’ is close to Vitis vinifera subsp. sylvestris and Vitis vinifera.
Vitis vinifera ‘Kyoho’ (Vitis vinifera × Vitis labrusca), a famous table grape cultivar, has strong anti-adversity and wide adaptability. Vitis vinifera ‘Kyoho’ has been cherished for their exquisite flavor, color, and health benefits. Recently, the findings about complete chloroplast genome sequence of different grapes such as Vitis vinifera grape Muscat Hamburg (Guo et al. Citation2020), Vitis davidii (Zhang et al. Citation2018) have been reported. However, the study about Vitis vinifera ‘Kyoho’ raised questions that needed more study.
The leaves of Vitis vinifera ‘Kyoho’ were collected from vineyard in Yanshi, Luoyang, China (N34°38′5.96′′, E112°42′21.39′′). The specimen of Vitis vinifera ‘Kyoho’ is stored in the Plant Specimen Museum of Henan University of Science and Technology under accession number GRAPEVINE2018052801. The CTAB method (Qu et al. Citation1996) was used for genomic DNA extracting. Then, it needs to pass the purity test by Qubit 2.0 and NanoDrop. Chloroplast genome sequenced from libraries by using Illumina X ten platform were used for assembly by FastQC (Version 0.11.5), Skewer (Version 0.2.2) (Jiang et al. Citation2014), seqkit (Version v0.9.0) (Shen et al. Citation2016), and ABYSS (Version 2.0.0) (Simpson et al. Citation2009). The CPGAVAS platform and OGDRAW platform were used for the chloroplast genome annotation and map construction, respectively. The accession number in GenBank for chloroplast genome sequences of Vitis vinifera ‘Kyoho’ is MN149615.
The complete chloroplast genome of Vitis vinifera ‘Kyoho’ is 160,928 bp in length. Its GC content is 37.4%. The complete chloroplast genome contains 89,148 bp large single-copy (LSC), 19,072 bp small single-copy (SSC) region, and 26,354 bp a pair of identical inverted repeat (IR) regions. Their GC contents are 35.3%, 31.7%, and 43.0%, respectively. A total of 113 genes in chloroplast genome of Vitis vinifera ‘Kyoho’ were annotated, including 80 protein-encoding genes, 29 tRNA genes, and 4 rRNA genes.
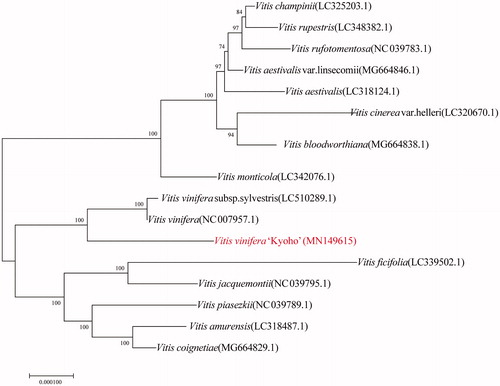
To understand the phylogenetic relationship and genetic diversity between Vitis vinifera “Kyoho” and other 15 Vitis species, the complete chloroplast genome sequences were collected and the neighbor-joining phylogenetic trees were constructed by MAGA software Version 5.2 (). NJ tree suggests that Vitis vinifera “Kyoho” is close to Vitis vinifera subsp. sylvestris (LC510289.1) and Vitis vinifera (NC 007957.1).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Guo DH, Li DM, Li H, Wang SP, Wang L, Niu JX, Ma C. 2020. The complete chloroplast genome sequence of Vitis vinifera Muscat Hamburg. Mitochondrial DNA Part B. 5(1):117–118.
- Jiang HS, Lei R, Ding SW, Zhu SF. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 15(1):182.
- Qu XP, Lu J, Lamikanra O. 1996. Genetic diversity in Muscadine and American bunch grapes based on randomly amplified polymorphic DNA (RAPD) analysis. JASHS. 121(6):1020–1023.
- Shen W, Le S, Li Y, Hu FQ. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLOS One. 11(10):e0163962.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.
- Zhang NB, Shen W, Wang R, Zhang JX. 2018. Complete sequence of Chinese wild Vitis davidii chloroplast DNA. Mitochondrial DNA Part B. 3(2):1021–1022.