Abstract
Vitis vinifera ‘Feng Zao’ is a bud mutation from V. vinifera ‘Kyoho’ which has better resistance and early ripening. The length of complete chloroplast genome sequence of V. vinifera ‘Feng Zao’ is 160,927 bp. The GC content is 37.4%. The chloroplast genome contains 89,147 bp large single-copy (LSC), 19,072 bp small single-copy (SSC), and 26,354 bp inverted repeat (IRs). Total 113 genes were annotated. The phylogenetic position of V. vinifera ‘Feng Zao’ is close to V. vinifera subsp. sylvestris.
Vitis vinifera ‘Feng Zao’, one kind of bud mutation of V. vinifera ‘Kyoho’, was found in 1998 (Guo and Zhang Citation2015). Vitis vinifera ‘Fengzao’ has a better resistance than V. vinifera ‘Kyoho’. Besides, V. vinifera ‘Fengzao’ ripens in early July which is almost 1 month earlier than V. vinifera ‘Kyoho’ (Guo et al. Citation2016). It has special rose aroma and suitable for field cultivation in the province of Henan, Shandong, Hebei, etc. (Guo and Zhang Citation2015).
Fresh leaf samples of V. vinifera ‘Feng Zao’ cultivated in vineyard located at Yanshi, Luoyang, China (N34°38′5.96′′, E112°42′21.39′′) were collected and used as material. Specimen of V. vinifera ‘Feng Zao’ is stored in the Plant Specimen Museum of Henan University of Science and Technology under the accession number GRAPEVINE2018052802. Genomic DNA was extracted using the CTAB method (Qu et al. Citation1996). Qubit 2.0 NanoDrop and agarose gel electrophoresis were used for purity and quality testing separately. Illumina X ten platform was used to sequence the DNA samples. The bioinformation software including FastQC version 0.11.5, Skewer version 0.2.2 (Jiang et al. Citation2014), seqkit version 0.9.0 (Shen et al. Citation2016), and ABYSS version 2.0.0 (Simpson et al. Citation2009) were used for chloroplast genome assembly of V. vinifera ‘Feng Zao’. The neighbor-joining phylogenetic tree construction, chloroplast genome annotation, and gene map completion were finished using MEGA version 5.2[Q] (Tamura et al. Citation2011), CPGAVAS platform, and OGDRAW platform, respectively. The accession number in GenBank of V. vinifera ‘Feng Zao’ is MN149614.
The complete chloroplast length of V. vinifera ‘Feng Zao’ is 160,927 bp. The total GC content is 37.4% and AT content is 62.6%. The GC content of 89,147 bp large single-copy (LSC) is 35.3%. The GC content of 19,072 bp small single-copy (SSC) is 31.74%. LSC and SSC are separated by 26,354 bp a pair of identical inverted repeats (IRs) region for which the GC content is 43.0%. The chloroplast genome of V. vinifera ‘Feng Zao’ contained 113 genes, including 80 protein-encoding genes, 29 tRNA genes, and 4 rRNA genes.
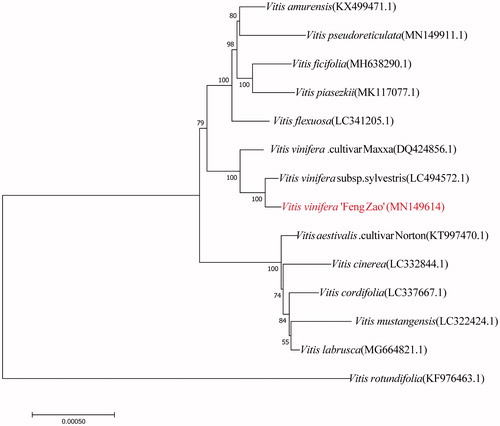
The complete chloroplast genome sequences of V. vinifera ‘Feng Zao’ and other 13 species of Vitaceae Vitis were collected and the neighbor-joining phylogenetic trees were constructed (). The result revealed the phylogenetic relationship and genetic diversity among these 14 species of Vitaceae Vitis. Vitis vinifera ‘Feng Zao’ is situated within the same branch with V. vinifera subsp. sylvestris (LC494572.1) and V. vinifera. cultivar Maxxa (DQ424856.1). The phylogenetic position of V. vinifera ‘Feng Zao’ in NJ tree is close to V. vinifera subsp. sylvestris (LC494572.1).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Guo DL, Xi FF, Yu YH, Zhang XY, Zhang GH, Zhong GY. 2016. Comparative RNA-Seq profiling of berry development between table grape ‘Kyoho’ and its early-ripening mutant ‘Feng Zao’. BMC Genom. 17(1):795.
- Guo DL, Zhang GH. 2015. A new early-ripening grape cultivar ‘Feng Zao’. Acta Hortic. 1082:153–156.
- Jiang HS, Lei R, Ding SW, Zhu SF. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf. 15(1):182.
- Qu XP, Lu J, Lamikanra O. 1996. Genetic diversity in Muscadine and American bunch grapes based on randomly amplified polymorphic DNA (RAPD) analysis. J Am Soc Hortic Sci. 121(6):1020–1023.
- Shen W, Le S, Li Y, Hu FQ. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 11(10):e0163962.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19(6):1117–1123.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.