Abstract
The complete plastid genome of Vanda subconcolor was assembled and analyzed. The genome size was 149,490 bp, containing a large single-copy region (LSC) of 85,691 bp, two inverted repeat regions (IRA and IRB) of 25,912 bp, and a small single-copy region (SSC) of 11,975 bp. The plastome contained 120 genes, including 74 protein-coding genes, 38 tRNA, and 8 rRNA. Phylogenomic analysis indicated that V. subconcolor is sister to V. brunnea.
Keywords:
Vanda R.Br. includes approximately 75 species (Gardiner and Cribb Citation2013; Zou et al. Citation2014, Citation2016), distributed from subtropical and tropical Asia to Australia (Queensland) and is one of the most important orchids in horticulture (Pridgeon et al. Citation2014). Vanda has been considered a ‘taxonomic black hole’ (Christenson Citation1987) and is one of the most taxonomically complicated groups in Aeridinae. Here, a complete plastid genome of Vanda, V. subconcolor, was assembled and annotated.
The total genomic DNA was extracted from fresh leaves using a modified Cetyltrimethylammonium Ammonium Bromide (CTAB) method (Doyle and Doyle Citation1987; Gardiner et al., Citation2013) and sequenced by the BGISEQ-500 platform. The samples were collected from Yulong Naxi Autonomous County (27°28′N, 100°26′E), Lijiang City, Yunnan Province of China and the voucher specimen deposited at Fujian Agriculture and Forestry University (specimen code MH Li or086). The clean reads were used to assemble the complete chloroplast genome by the GetOrganelle pipe-line (Jin et al. Citation2018), with the chloroplast genome of Vanda brunnea (GenBank accession MK442937) as the reference sequences. The assembled chloroplast genome was annotated using the Geneious R11.15 (Kearse et al. Citation2012).
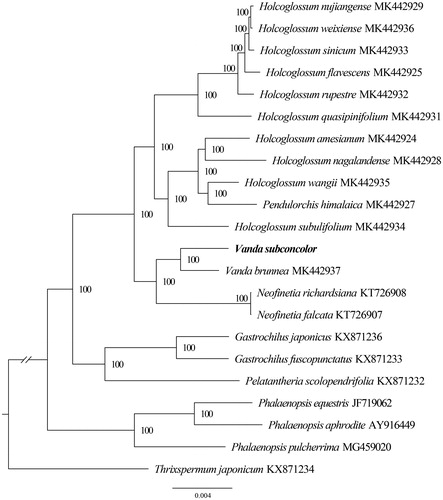
The complete plastid genome of V. subconcolor (GenBank accession MT180955) is 149,490 bp in length, containing a large single copy (LSC) region of 85,691 bp, two inverted repeat regions (IRA and IRB) of 25,912 bp, and a small single-copy region (SSC) of 11,975 bp. The new complete plastid genome contained 120 genes, including 74 protein-coding genes, 8 rRNA genes, and 38 tRNA genes. The overall GC content of the chloroplast genome was 36.64% (LSC, 33.96%; IR, 43.08%; SSC, 27.91%). To confirm the phylogenetic position of V. subconcolor, 22 representative species of Aeridinae were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and phylogenetic tree constructed by RAxML (Stamatakis Citation2014) (). The ML tree showed that V. subconcolor is sister to V. brunnea with strong support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Christenson EA. 1987. An infrageneric classification of Holcoglossum Schltr. with a key to the genera of the Aerides-Vanda alliance. Notes R Bot Garden Edinburgh. 44:249–256.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gardiner LM, Kocyan A, Motes M, Roberts DL, Emerson BC. 2013. Molecular phylogenetics of Vanda and related genera (Orchidaceae). Bot J Linn Soc. 173(4):549–572.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. 2014. Genera Orchidacearum, volume 6: Epidendroideae (part three). New York (NY): Oxford University Press; p. 554.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zou LH, Liu ZJ, Huang JX. 2014. Vanda malipoensis, a new species of Vanda (Orchidaceae: Epidendroideae; Vandeae) from China: evidence from morphological and molecular phylogenetic analyses. Phytotaxa. 186(2):87–96.
- Zou LH, Zou LH, Wu XY, Lin M, Chen LJ, Liu ZJ. 2016. Vanda funingensis, a new species of Orchidaceae (Epidedroideae; Vandeae; Aeridinae) from China: evidence from morphology and DNA. Phytotaxa. 260(1):1–13.