Abstract
In this study, the whole chloroplast genome of Ulmus glabra ‘Pendula’ was sequenced, assembled, annotated and characterized for the first time. The results showed that the total length of the chloroplast genome of U. glabra ‘Pendula’ was 159,305 bp and the GC content was 35.58%. It had a circular tetrad structure of the chloroplast genome of typical angiosperms, including a large single-copy (LSC) with length of 87,916 bp, a small single-copy (SSC) with length of 18,693 bp, and a pair of inverted repeat sequence (IRs) of 26,348 bp. A total of 131 genes were identified, including 86 protein coding genes, 37 tRNA genes, and 8 rRNA genes. Based on the whole chloroplast genome sequence, the phylogenetic trees of 14 plants in U. glabra ‘Pendula’ and NCBI database were constructed, and the phylogenetic position of U. glabra ‘Pendula’ in Ulmaceae was identified. The results provided important information for the identification of genetic relationship of U. glabra ‘Pendula’, and will also promote the functional analysis of genes related to agronomic traits controlled by the chloroplast genome of U. glabra ‘Pendula’.
The chloroplast genome is an organelle genome independent of the nuclear genome. It has a separate transcription and transport system, which mainly encodes genes related to photosynthesis and chloroplast-specific components (Wang et al. Citation2019), and involves multiple metabolic pathways (Lin et al. Citation2014), such as sugar synthesis, starch storage, and pigment synthesis. Most of the chloroplast genomes are circular double-stranded structures, ranging in size from 120 to 190 kb, with relatively conservative sequence characteristics (Palmer Citation1985; Yan and Zhu Citation2004) and individual gene variation (Hollingsworth et al. Citation2009). Therefore, by determining the chloroplast genome of U. glabra ‘Pendula’ and based on the phylogenetic analysis of Ulmaceae, the phylogenetic relationship between U. glabra ‘Pendula’ and Ulmaceae can be identified, which provides a powerful supplement for the taxonomic study of Ulmaceae.
Ulmus glabra ‘Pendula’ is a cultivated variety of Ulmus glabra f.cornuta, which has the characteristics of drooping branches, large leaves and grow very close together. The experimental materials were planted in Hebei Academy of Forestry and Grassland, Shijiazhuang, China (114°28′12″E, 38°08′23″N), and the file number is HAFG22Uugl. Ulmus glabra ‘Pendula’ DNA was extracted, chloroplast genome was sequenced, assembled and annotated by Beijing Zhongxing Bomai Technology Co., Ltd, and the sequencing platform was Illumina NovaSep. The DNA is now stored in Hebei Engineering Research Center for Trees Varieties.
The chloroplast genome of U. glabra “Pendula” was sequenced with high quality, and a complete circular chloroplast genome sequence (uploaded to NCBI with the number of MT160186) was obtained after assembly and splicing. The total length of U. glabra “Pendula” chloroplast genome sequence is 159,305 bp, which has a typical circular tetrad structure of angiosperm chloroplast genome, including a LSC region of 87,916 bp, a SSC region of 18,693 bp, a pair of IRs regions 26,348 bp, respectively. The total GC content of the genome was 35.58%. A total of 131 genes were annotated, including 86 protein coding genes, 37 tRNA genes, and 8 rRNA genes.
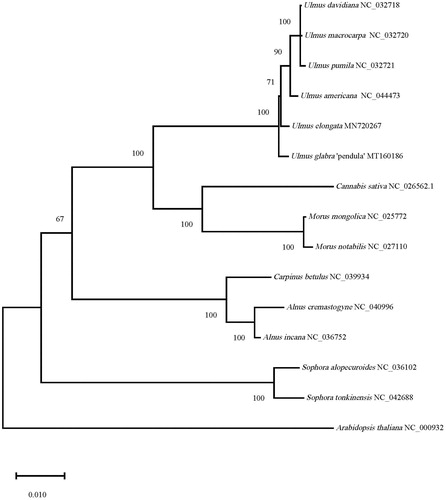
Phylogenetic trees were constructed based on U. glabra “Pendula” and 13 closely related chloroplast genomic sequences downloaded from NCBI database, including five Ulmus species, two Morus species, one Cannabis specie, two Sophora species, two Alnus species, one Carpinus specie, in addition, the outgroup is Arabidopsis thaliana. The phylogenetic tree was constructed by the maximum-likelihood method and repeated 1000 times, and the result was reliable. The results showed that (), U. glabra ‘Pendula’ and five varieties of Ulmus were clustered into one branch and differentiated first, and then differentiated into the U. elongate of the Sect. Chaetoptelea, the U. americana of the Sect. Blepharocarpa and the U. davidiana, U. macrocarpa, U. pumila of the Sect. Ulmus. The result defines the phylogenetic position of U. glabra ‘Pendula’ at the molecular level for the first time, further improves the chloroplast genome information in Ulmus, and provides a theoretical basis for further research on the exploitation and utilization, origin and evolution of Ulmus resources.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, et al. 2009. A DNA barcode for land plants. Proc Natl Acad Sci. 106(31):12794–12797.
- Lin Z, Wang Y, Fu F, Ye C, Fan L. 2014. Assembly and phylogenetic analysis of Dongxiang wild rice chloroplast genome. J Zhejiang Univ (Agri Life Sci). 40(4):397–403.
- Palmer JD. 1985. Comparative organization of chloroplast genome. Annu Rev Genet. 19(1):325–354.
- Wang X, Zheng Q, Luo Y, Bai Z, Li W, Li J, Cai Y. 2019. Characterization of the complete chloroplast genome of the ‘Huishui JinJu’. J Fruit Sci. 36(3):257–265.
- Yan A, Zhu D. 2004. The application of chloroplast genome in the studies of systematics and bioengineering. Chin J Cell Biol. 26(2):153–156.