Abstract
The complete mitochondrial genome of Medinilla magnifica, an economically valuable species in the family Melastomataceae was reported in this study. The genome size is 377,864 bp in length and the GC content is 43.91%. A total of 98 genes were annotated, including 71 protein-coding genes, 24 tRNA genes and 3 rRNA genes. Phylogenetic analysis revealed a close relationship between M. magnifica with Eucalyptus grandis. This is the first complete mitochondrial genome for M. magnifica that would be useful for providing basic genetic information for this important species.
Medinilla (Myrtales, Melastomataceae) is a remarkable genus with highly ornamental value because of their broad leaves and showy flowers. Medinilla magnifica is a perennial evergreen shrub in this genus and originated in Philippines (Robil and Tolentino Citation2015). It is called the Lotus Lantern in China because its flower is like the traditional Chinese lantern (Yang et al. Citation2009). In this study, we assembled and characterized the complete mitochondrial genome of M. magnifica for the first time in order to provide basic genetic information for this important species. The annotated mitochondrial genome sequence was submitted to GenBank under the accession number of MT043351.
The plant material of M. magnifica was collected from Liaoning Academy of Agricultural Sciences (N41°48’33”, E123°34’53”), Shenyang, China. The specimen was stored with the archival number of MELA_MED_MAG_01 at Institute of Floriculture of Liaoning Academy of Agricultural Sciences. Total genomic DNA was extracted and sequenced on the Illumina Miseq sequencing platform by Shanghai Personal Biotechnology Co. Ltd, China. Clean pair-end reads were filtered and assembled using SPAdes v.3.11.0 software (Bankevich et al. Citation2012) to a complete mitochondrial genome. Genes were annotated by homology alignments and de novo prediction.
The complete mitochondrial genome of M. magnifica is 377,864 bp in length with an overall GC content of 43.91%. A total of 98 genes were annotated, including 71 protein-coding genes, 3 rRNA genes and 24 tRNA genes. There are two protein-coding genes (rps10 and ccmFC) having one intron, two (nad4 and nad5) having three introns, and three (nad1, nad2 and nad7) having four introns. Among the tRNA genes, trnG-GCC is duplicated and trnM-CAT is in tuplicate along the mitochondrial genome.
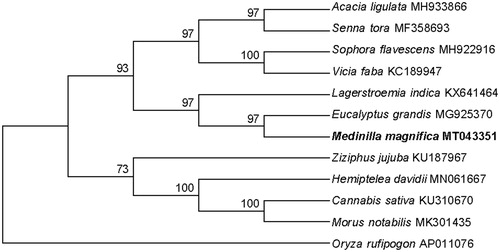
The phylogenetic relationships of M. magnifica and eleven other plant mitochondrial genomes were evaluated based on protein-coding genes. The phylogenetic tree was constructed using maximum-likelihood analysis with MEGA 7.0 (Kumar et al. Citation2016). The result showed that M. magnifica is closely related to Eucalyptus grandis (Myrtales, Myrtaceae). Our mitochondrial genome is the first submitted Melastomataceae mitochondrial genome and the third of Myrtales in the GenBank database (Pinard et al. Citation2019), and will provide valuable genetic information for M. magnifica species.
Disclosure statement
The authors declare no competing financial interest. The authors alone are responsible for the content and writing of the paper.
Figure 1. Phylogenetic relationship of Medinilla magnifica and eleven related species based on mitochondrial genome sequences. The phylogenetic tree was constructed using maximum-likelihood method based on the protein-coding genes, with Oryza rufipogon as the outgroup. The branch support was determined by computing 1000 non-parametric bootstrap replicates.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Pinard D, Myburg AA, Mizrachi E. 2019. The plastid and mitochondrial genomes of Eucalyptus grandis. BMC Genomics. 20(1):132.
- Robil JLM, Tolentino VS. 2015. Histological localization of tannins at different developmental stages of vegetative and reproductive organs in Medinilla magnifica (Melastomataceae). Flora. 217:82–89.
- Yang TS, Ke SF, Zhao L, Jiang L. 2009. Study on cottage propagation technique in Medinilla magnifica. Tianjin Agr Sci. 15(4):11–12.
