Abstract
The complete mitochondrial genome of Melanostoma scalare was sequenced and reported here. The circle genome of the syrphid fly is 16,380 bp in length. There are 38 sequence elements including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. The order of all elements was the same with that of Eristalia cerealis. With 2 species from Muscidae and Drosophilidae as outgroups, phylogenetic relationships of 8 Syrphidae species based on mitogenomes were in complete agreement with their taxonomic relationships based on morphological characteristics. Our result will provide more fundamental data to the development of the molecular systematics of Syrphidae.
Melanostoma scalare (Fabricius, 1794), belongs to the Eristalini tribe in family Syrphidae, one of the most diverse families in Diptera, including about 6000 described species worldwide (Thompson Citation2013). Syrphidae insects are commonly known as hoverflies, flower flies, or sunflies because of their peculiar behavior of hovering in sun over the flowers to feed on pollen and nector (Khan Citation2017). Syrphidae adults have been regarded as important pollinators all along. Melanostoma scalare can be easily distinguish from other Melanostoma species by its pilose arista, densely dusted face, and long abdomen (Stahls and Haarto Citation2014). Here, we sequenced the complete mitogenome of M. scalare to provide more fundamental data to the development of the DNA barcoding system.
In this study, M. scalare adults were collected from Xining Botanical Garden, Qinghai, China (36°37′36″N, 101°44′58″E) in September 2019 and kept in the Insect Collection of the Entomology Lab, College of Agriculture and Animal Husbandry, Qinghai University, Xining, China (accession number: YJY-2019-SYY003). After genomic DNA extraction, genomic sequencing was performed on the Illumina HiSeq Platform (Illumina, San Diego, CA) with a read length of 150 bp. The software MITObim (Hahn et al. Citation2013) was employed to assemble the mitogenome. The assembled sequence was then annotated using the web server MITOS (Bernt et al. Citation2013).
A circularized DNA assembly 16,380 bp in length was harvested and deposited in the GenBank with the accession number of MT185683. Similar to the reported Syrphidae mitogenomes, 37 genes are recognized in this mitogenome: 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes, as well as a control region (D-loop). Of all 38 sequence elements, 4 PCGs, 8 tRNA genes, and 2 rRNA genes are located on the light strand, while others are on the heavy strand. The order of all elements distributed on the sequence is consistent with that of Eristalia cerealis (Yan et al. Citation2020).
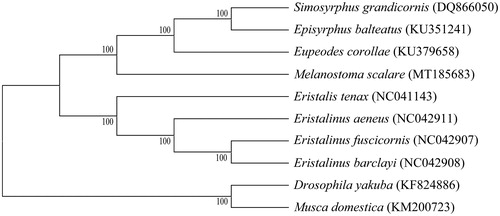
Based on mitogenomes assembled here or downloaded from GenBank, phylogenetic relationships of 8 Syrphidae species with 2 species from Muscidae and Drosophilidae as outgroups were resolved by means of neighbor-joining (). After aligned using MAFFT (Katoh and Standley Citation2013), the neighbor-joining tree was built using MEGA7 (Kumar et al. Citation2016) with bootstrap set to 1000. Phylogenetic relationships indicated by the phylogenetic tree were in complete agreement with their taxonomic relationships based on morphological characteristics.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khan AA. 2017. Effect of insecticides on biodiversity of Aphidophagous syrphid flies in fruit ecosystem of Kashmir. J Entomol Zool Stud. 5(4):117–125.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Stahls G, Haarto A. 2014. When mtDNA COI is misleading: congruent signal of ITS2 molecular marker and morphology for North European Melanostoma Schiner, 1860 (Diptera, Syrphidae). ZooKeys. 431:93–134.
- Thompson FC. 2013. Family Syrphidae. In: Thompson FC, Pape T, editors. Systema Dipterorum, Version 1.5. [accessed 2015 Aug 15]. Online at http://www.diptera.org.
- Yan J, Feng S, Song P, Li Y, Li W, Liu D, Ju X. 2020. Characterization and phylogenetic analysis of the complete mitochondrial genome of Eristalia cerealis (Diptera: Syrphidae). Mitochondrial DNA Part B. 5(1):1005–1006.