Abstract
Coriaria nepalensis Wall. is a toxic Chinese herbal medicine. In this study, the complete chloroplast (cp) genome of C. nepalensis was first reported. The length of the large single-copy (LSC) region was 158,499 bp, the small single-copy (SSC) region was 18,411 bp, and the inverted repeats (IRs) region were 26,552 bp, with 37.76% overall GC content. There were 132 genes in total, including 86 mRNA, 38 tRNA, and 8 rRNA. Phylogenetic analysis showed that C. nepalensis was closely related to Corynocarpus laevigatus.
Coriaria nepalensis Wall. is a shrub belonging to single-genus of Coriaria (Coriariaceae family), and mainly distributed in the southern and southwestern of China. It is commonly used for the treatment of numbness, toothache, traumatic injury, and acute conjunctivitis in traditional Bai folk medicine (Jiang and Duan Citation2014), which are toxic to many animals (Wang et al. Citation2016). Previous studies on C. nepalensis mainly focused on chemical components and biological characteristics (Wang et al. Citation2016; Zhao et al. Citation2018). However, the complete chloroplast (cp) genome sequence of C. nepalensis has not been reported so far. Herein, we assembled and characterized the cp genome of C. nepalensis for the first time. Besides, its phylogenetic relationship with several plant species was investigated, which will provide significant genomic information to study the evolution of the Coriariaceae.
Fresh leaves of C. nepalensis were collected from the Cangshan mountain (25°67′30″N, 100°15′72″E) of Dali counties, Yunnan province, China. The voucher specimen was deposited in the herbarium of Dali University (LJ2020010506). Total DNA was extracted using the DNeasy plant mini kit (QIAGEN), and sequenced by Illumina NovaSeq system (Illumina, San Diego, CA, USA). About 8.05 Gb of raw data with 53,700,486 paired-end reads were assembled by NOVOPlasty (Dierckxsens et al. Citation2017; Park et al. Citation2019), and the assembled cp genome was annotated by GeSeq with default sets (Michael et al. Citation2017; Qian et al. Citation2020). The annotated cp genome of C. nepalensis was submitted to the GenBank with the accession number of MT106774.
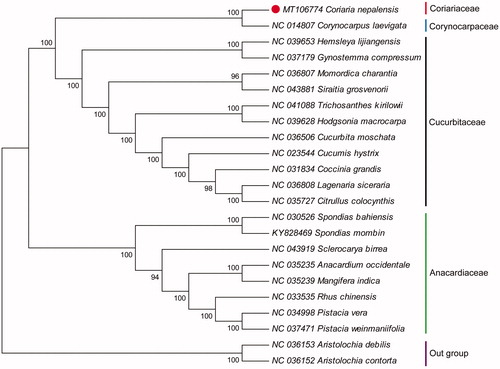
The cp genome of C. nepalensis was 158,499 bp in length, with a typical quadripartite structure of large single-copy (LSC) region of 86,984 bp, and a small single-copy (SSC) region of 18,411 bp, separated by a pair of inverted repeats (IRs) region of 26,552 bp. It contained 132 genes, including 86 protein-coding genes, 38 tRNA genes, and 8 rRNA. The overall GC content was 36.76%. To confirm the phylogenetic relationship of C. nepalensis, a phylogenetic analysis was performed based on 20 cp genomes. Meanwhile, two species from the Aristolochiaceae (Aristolochia debilis and Aristolochia contorta) were used as outgroups. The sequence was aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and then a neighbour-joining (NJ) tree was constructed using MEGA 6.0 (Tamura et al. Citation2013) with 1000 bootstrap replicates. The result showed that C. nepalensis was closely related to Corynocarpus laevigatus (). In total, this study reports the first cp genome of species in Coriariaceae which would be beneficial to potential studies on phylogenetics of the genus and related group in Coriariaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 4(45):e18.
- Jiang B, Duan BZ. 2014. Bai’s traditional plant medicine. Beijing: China Press of Traditional Chinese Medicine.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Michael T, Pascal L, Tommaso P, Elena SU, Axel F, Ralph B, Stephan G. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Park J, Choi YG, Yun N, Xi H, Min J, Kim Y, Oh S-H. 2019. The complete chloroplast genome sequence of Viburnum erosum (Adoxaceae). Mitochondrial DNA B. 4(2):3278–3279.
- Qian J, Du ZF, Jiang Y, Duan BZ. 2020. The complete chloroplast genome sequence of Althaea rosea (L.) Cavan. (Malvaceae) and its phylogenetic analysis. Mitochondrial DNA B. 5(2):1433–1434.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol E. 30(12):2725–2729.
- Wang YY, Tian JM, Zhang CC, Luo B, Gao JM. 2016. Picrotoxane sesquiterpene glycosides and a coumarin derivative from Coriaria nepalensis and their neurotrophic activity. Molecules. 21(10):1344.
- Zhao F, Liu YB, Ma SG, Yu DQ, Yu SS. 2018. New compounds from the roots of Coriaria nepalensis. Chinese Chem Lett. 29(3):467–470.