Abstract
Bidens frondosa is an annual invasive alien species of Asteraceae. The complete plastid genome of B. frondosa was determined and analyzed in this study. The genome was 150,490 bp in length, including a large single-copy region (LSC) of 83,500 bp, a small single-copy region (SSC) of 17,628 bp, and a pair of inverted repeat regions (IR) of 24,681 bp each. The plastid genome encodes a set of 135 genes, which comprised 87 protein-coding genes, 8 ribosomal RNA genes, and 40 transfer RNA genes.
Bidens frondosa L. is an invasive herbaceous annual species originated from North America. It was intentionally introduced into many countries as herbs and horticultural plants (Danuso et al. Citation2012). Moreover, its seed can also disperse into new areas by water, animals, or grain transportation (Šumberová et al. Citation2004). There is evidence that this species can displace native species of Bidens or interbreed with them and replace them with hybrids (Vasilyeva and Papchenkov Citation2011). It also can inhibit seed germination of Plantago virginica and Ageratum conyzoides through allelopathy (Yan et al. Citation2012). The spreading and colonization of B. frondosa can lead to biodiversity loss and agricultural production reduce (Yan et al. Citation2012). However, the essential oil of B. frondosa contains antioxidant and antimicrobial properties, and this species can be used as medicinal plants (Rahman et al. Citation2011).
Fresh leaves of B. frondosa were collected from the Damingshan National Nature Reserve in Nanning city, Guangxi Zhuang Autonomous Region of China (108°26′21″E, 23°30′47″N), for total genomic DNA extraction. The voucher specimen was preserved at the Herbarium of Shiwandashan National Nature Reserve (SWDS, accession number DMS190803). High-throughput DNA sequencing was conducted on the Illumina Nova-seq Sequencing System (Illumina, CA, USA), and sequenced by Jierui Biotch (Guangzhou, China). 7.10 Gb clean data were obtained after removing low-quality reads and adaptor sequences. We assembled the complete cp genome of B. frondosa using GetOrganelle pipeline (available online: https://github.com/Kinggerm/GetOrganelle), (Jin et al. Citation2018). The cp genome annotation was accomplished using Geseq (available online: https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017) coupled with manual check and adjustment. Finally, the validated complete chloroplast genome sequence was submitted to GenBank with the accession number MT178455.
The complete chloroplast genome of B. frondosa (MT178455) is 150,490 bp in size, containing a large single-copy (LSC) region of 83,500 bp, a small single-copy (SSC) region of 17,628 bp and a pair of inverted repeat (IR) regions of 24,681 bp. The overall GC-content of the whole plastome is 37.54%. This chloroplast genome harbors 135 functional genes, including 87 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. Among all of these genes, 21 are involved in photosynthesis. 2 genes (clpP and ycf3) contain double introns, while 12 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpoC1, rps12, rps16, trnA-UGC, trnI-GAU, trnK-UUU) harbor a single intron.
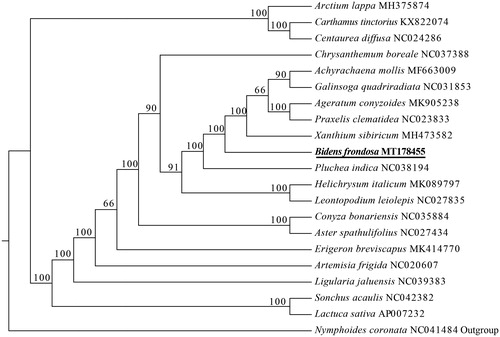
In order to reveal phylogenetic locations of B. frondosa and other members of the Asteraceae, maximum likelihood (ML) analysis was performed based on 20 complete chloroplast genomes of Asteraceae and one species of Menyanthaceae as outgroup. All of them were downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and the ML phylogenetic tree was constructed by RaxML (Stamatakis Citation2014). The phylogenetic tree showed that B. frondosa was a sister clade to Achyrachaena mollis – Xanthium sibiricum clade with strong support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Danuso F, Zanin G, Sartorato I. 2012. A modelling approach for evaluating phenology and adaptation of two congeneric weeds (Bidens frondosa and Bidens tripartita). Ecol Modell. 243:33–41.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Yi T-S, Li D-Z. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv.:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Rahman A, Bajpai VK, Dung NT, Kang SC. 2011. Antibacterial and antioxidant activities of the essential oil and methanol extracts of Bidens frondosa Linn. Int J Food Sci Technol. 46(6):1238–1244.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Šumberová K, Tzonev R, Vladimirov V. 2004. Bidens frondosa (Asteraceae) – a new alien species for the Bulgarian flora. Phytologia Balcanica. 10:179–181.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Vasilyeva N, Papchenkov V. 2011. Mechanisms of influence of invasive Bidens frondosa L. on indigenous Bidens species. Russ J Biol Invasions. 2(2-3):81–85.
- Yan X, Zeng J, Zhou B, Wang N, Xiang H, Kang Y. 2012. Allelopathic potential of the extracts from alien invasive plant Bidens frondosa. J Yangzhou Univ Agri Life Sci Ed. 33:88–94.