Abstract
Members of the Vaccinium genus (Ericaceae family) have excellent medicinal and nutritional properties. In this study, we sequenced the complete chloroplast (cp) genomes of three species, Vaccinium bracteatum Thunb., V. vitis-idaea L., and V. uliginosum L., to investigate their phylogenetic relationships within the Ericaceae. The chloroplast genomes of these species are 174,404 bp, 173,967 bp, and 173,356 bp, respectively. These genomes have a typical quadripartite structure, with an LSC region (106,565 bp, 106,013 bp, and 105,856 bp) and an SSC region (2979 bp, 3518 bp, and 3146 bp) separated by a pair of IRs (32,430 bp, 32,218 bp, and 32,177 bp, respectively). These cp genomes are composed of 139 genes, including 93 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Maximum likelihood phylogenetic analysis indicated that V. bracteatum is most closely related to V. oldhamii. Our findings should facilitate further evolutionary and genetic studies.
The genus Vaccinium of the Ericaceae family comprises more than 450 species across Europe, North America, Central America, Central and South East Africa, Madagascar, and Asia. Many Vaccinium species are popular berry crops, such as blueberry (V. ashei J.M. Reade, V. angustifolium Aiton, V. corymbosum L.), bilberry (V. myrtillus L.), cranberry (V. macrocarpon Aiton, V. oxycoccus L.), huckleberry (V. ovatum Pursh, V. parvifolium Sm.), sea blueberry (V. bracteatum Thunb.), oldham blueberry (V. oldhamii Miq.), bog blueberry (V. uliginosum L.), and lingonberry (V. vitis-idaea L.). Among these, V. bracteatum, V. oldhamii, V. uliginosum, and V. vitis-idaea are widely distributed in Korea and are commonly consumed as foods (Luby et al. Citation1991; Forney and Kalt Citation2011). To date, most studies have focused on the commercial properties of these berries including their chemical compositions. However, there are no reports of the complete chloroplast (cp) genome sequences of any Vaccinium species except V. oldhamii. Here, we report the complete cp genome sequences of three Vaccinium species, V. bracteatum, V. vitis-idaea, and V. uliginosum, based on Illumina pair-end sequencing data, which should facilitate evolutionary and genetic research.
Leaf samples from V. bracteatum, V. vitis-idaea, and V. uliginosum were collected from Jindo-gun (N34°22′34.6″, E126°17′19.8″) in Jeollanam-do (province), Hongcheon-gun (N37°47′31.5″, E128°15′11.8″) in Gangwon-do (province), and Seogwipo-si (N33°21′36.7″, E126°31′44.5″) in Jeju-do (island), Korea, respectively. The specimens were deposited at the National Institute of Biological Resources (NIBR), Korea (specimen codes NIBR0000424891, NIBRVP0000460961, and NIBR0000424897, respectively).
Total genomic DNA was extracted from fresh leaves using the CTAB method. Whole genome resequencing of the cp genomes was performed on the Illumina HiSeq X Ten platform by SEEDERS Inc. (Daejeon, Korea). Raw reads were filtered using Dynamic Trim and Length Short in the SolexaQA (ver.1.13) package (Cox et al. Citation2010).
Clean reads were aligned to the cp genome of V. oldhamii (Genbank Accession No. NC_042713). Filtered reads were assembled into contigs using NOVOPlasty v 2.7.1 (Dierchxsens et al. Citation2017) and Fast-Plast v1.2.8 software (Mckain et al. Citation2018). Physical maps of the newly assembled cp genomes were generated using OGDRAW v 1.3.1 (Stephan Citation2019). The validated complete cp genome sequences were submitted to GenBank under accession numbers LC521967, LC521969, and LC521968, respectively.
The total lengths of the cp genome sequences of V. bracteatum, V. vitis-idaea, and V. uliginosum were 174,404 bp, 173,967 bp, and 173,356 bp, respectively. In the cp genome of V. bracteatum, a pair of 32,430-bp inverted repeats (IRs) was separated by a small single-copy (SSC) region of 2979 bp and a large single-copy (LSC) region of 106,565 bp. Vaccinium vitis-idaea contained a 106,013-bp LSC region, a 3518-bp SSC region, and a 32,218-bp pair of IRs. Vaccinium uliginosum contained a 105,856-bp LSC region, a 3146-bp SSC region, and a 32,177-bp pair of IRs. Each cp genome sequence contained 139 genes, including 93 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC content of the whole plastome of each species was 36.8%.
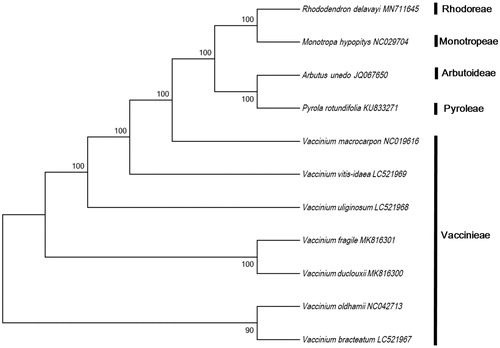
To investigate the phylogeny of V. bracteatum, V. vitis-idaea, and V. uliginosum, the complete cp genome sequences of nine Ericaceae species (Vaccinium fragile, Vaccinium duclouxii, Vaccinium oldhamii, Vaccinium macrocarpon, Rhododendron delavayi, Rhododendron pulchrum, Arbutus unedo, Pyrola rotundifolia, and Monotropa hypopitys) were aligned using ClustalW (Thompson et al. Citation1994). The data were downloaded from NCBI GenBank. A maximum-likelihood (ML) tree was constructed using MEGA 7.0 (Kumar et al. Citation2016) with 1000 bootstrap replicates.
ML tree analysis indicated that V. bracteatum shares a closer relationship with V. oldhamii than with the other species, with 90% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinf. 11(1):485.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4): e18–e18.
- Forney CF, Kalt W. 2011. Blueberry and cranberry. Health-promoting properties of fruit and vegetables. Wallingford, UK: CABI.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Luby JJ, Ballington JR, Draper AD, Pliszka K, Austin ME. 1991. Blueberries and cranberries (Vaccinium). Acta Hortic. 290(290):393–458.
- McKain MR, Wilson MC, Kellogg EA. 2018. Fast-Plast: a rapid de novo assembly pipeline for whole chloroplast genomes. Git-Hub. 973887. doi:10.5281/zenodo,2018,973887.
- Stephan G. 2019. Lehwark Pascal, Bock Ralph. Organellar Genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 22(22):4673–4680.