Abstract
Morinda parvifolia is a traditional medicinal plant of the family Rubiaceae. In the current study, the complete chloroplast genome of M. parvifolia was sequenced and reported for the first time. The genome is 153,069 bp in total length, containing two inverted repeats (IR) regions of 25822 bp which were separated by a large single copy (LSC) region of 83813 bp and a small single copy (SSC) region of 17612 bp. A total of 131 gene species were annotated, including 86 protein-coding genes,37 tRNA genes, and 8 rRNA genes. The overall GC content of the genome is 38.07%. The phylogenetic analysis based on the complete chloroplast genome reveals that M. parvifolia is closely related to Morinda officinalis. This study enriches the genetic information of M. parvifolia as well as for the phylogenetic studies for Rubiaceae.
Morinda parvifolia Bartl. ex DC. is a perennial liana or creeping shrub of the family Rubiaceae, which grows at sparse forests or forest margins in southeastern Asia, including the south of China, Vietnam, and Philippines (Chen and Taylor Citation2011). It is used as a traditional medicine in the treatment of tracheitis, coughing and dyspepsia (Su et al. Citation2019), which contains the compounds of quinones, flavonoid glycosides, and terpenoids, etc (Chang et al. Citation1982; Chang and Lee Citation1984; Kang et al. Citation2016). However, recently, overexploitation has adversely affected its natural distribution, population, and habitat. Meanwhile, M. parvifolia might be the first case showing the possibility that androdioecy could be a mid-stage in the pathway of dioecy evolving from stigma-height dimorphism (Liu et al. Citation2012). To date, there have been no studies on the genome of M. parvifolia. The chloroplasts (cp) genome has a maternal inheritance and conserved structure, which has been used for investigating evolutionary and phylogenetic relationships of plants (Wang et al. Citation2018). Therefore, in the present study, we reported and characterized the complete chloroplast (cp) genome sequence of M. parvifolia by next-generation sequencing technology. Additionally, a phylogenomic analysis of this species and the relatives was also presented.
Fresh leaves of M. parvifolia were collected from Huboliao National Nature Reserve of Fujian province, China (N24.89°, E117.24°). The specimens (MS20181121) were deposited in the herbarium of Minnan Normal University. Total genomic DNA was extracted from fresh leaves using a modified CTAB method (Murray and Thompson Citation1980) and was used further to construct the library with an average length of 350 bp by using the Nextera XT DNA Library Preparation Kit. The high-throughput sequencing was performed on the Illumina Novaseq platform, and the average length of the generated reads was 150 bp. The Illumina raw sequence reads were edited using the NGS QC Tool Kit v2.3.3 (Ge et al. Citation2018), and high-quality reads were assembled into contigs using the de novo assembler SPAdes3.11.0 (Bankevich et al. Citation2012). The complete chloroplast sequence was annotated by Plann software (Huang and Cronk Citation2015) and has been deposited in GeneBank with the accession number (MT157221).
The complete chloroplast genome of M. parvifolia has been retrieved with an average sequencing depth of 1047X and is 153,069 bp in total length. It had a typical quadripartite structure, containing a large single copy (LSC) region of 83813 bp and a small single copy (SSC) region of 17612 bp which were separated by two inverted repeats (IRA and IRB) regions of 25822 bp. A total of 131gene species were annotated, including 86 protein-coding genes,37 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA), respectively. Among them, fifteen genes (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA) contain a single intron, and two genes (clpP, ycf3) contain two introns. Besides, one PCG (rps12) occurred in trans-splicing which is divided into three parts including the first part within LSC and the two exons within IR regions, respectively. Most of the genes appear to be single copy except eighteen genes which have two copies including seven protein-coding genes (rps12,rpl23, rpl2, rps7, ndhB, ycf2, and ycf1), four rRNAs (rrn4.5, rrn5, rrn16, and rrn23) and seven tRNAs (trnL-CAA, trnV-GAC, trnA-UGC, trnI-GAU, trnN-GUU, trnR-ACG, and trnI-CAU). The base composition of the cp genome for M. parvifolia is uneven (30.64% A,19.38%C,18.69%G and 31.28%T) with an overall GC content of 38.07%, which is close to those of its confamilial plants, for example, Hedyotis ovata (MK203877) (Zhang et al. Citation2019) and Morinda officinalis (NC_028009) (Zhang et al. Citation2016).
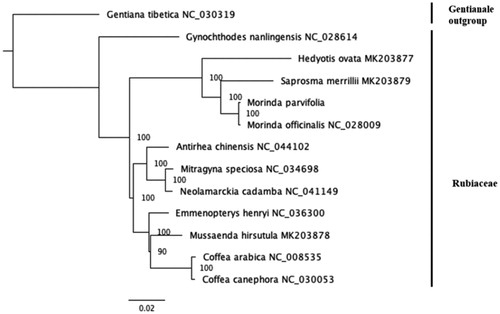
The obtained complete chloroplast genome sequence of M. parvifolia together with the complete chloroplast genomes of 12 taxa downloaded from NCBI GenBank were applied to perform the phylogenetic analysis. We used RAxML v8.2.9 software (Stamatakis Citation2014), with 1000 bootstrap replicates to reconstruct a maximum-likelihood phylogenetic tree, using Gentiana tibetica as an outgroup. The phylogenetic result has shown that M. parvifolia is closely related to M. officinalis ().
Disclosure statement
The authors confirm this article content has no conflict of interest, and all the authors are responsible for the content of this article.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chang P, Lee K-H. 1984. Cytotoxic antileukemic anthraquinones from Morinda parvifolia. Phytochemistry. 23(8):1733–1736.
- Chang P, Lee K-H, Shingu T, Hirayama T, Hall IH, Huang H-C. 1982. Antitumor agents 50. morindaparvin-A, a new antileukemic anthraquinone, and alizarin-1-Methyl ether from morinda parvifolia, and the antileukemic activity of the related derivatives. J Nat Prod. 45(2):206–210.
- Chen T, Taylor CM. 2011. Flora of China. Beijing: Science Press. Vol. 19; p. 220–227.
- Ge J, Cai L, Bi G-Q, Chen G, Sun W. 2018. Characterization of the complete chloroplast genomes of Buddleja colvilei and B. sessilifolia: implications for the Taxonomy of Buddleja L. Molecules. 23(6):1248.
- Huang DI, Cronk Q. 2015. Plann: A command‐line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kang J, Zhang P, Gao Z, Zhang J, Yan Z, Wang H, Chen R. 2016. Naphthohydroquinones, naphthoquinones, anthraquinones, and a naphthohydroquinone dimer isolated from the aerial parts of Morinda parvifolia and their cytotoxic effects through up-regulation of p53. Phytochemistry. 130:144–151.
- Liu Y, Luo Z, Wu X, Bai X, Zhang D. 2012. Functional dioecy in Morinda parvifolia (Rubiaceae), a species with stigma-height dimorphism. Plant Syst Evol. 298(4):775–785.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 8(19):4321–4326.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su X, Li L, Sun H, Zhang F, Li C, Li F, Wang H, Li B, Chen R, Kang J. 2019. Eight new glycosides with hepatoprotective activity isolated from the aerial parts of Morinda parvifolia. Bioorg Chem. 87:867–875.
- Wang J, Li C, Yan C, Zhao X, Shan S. 2018. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PeerJ. 6:e5349.
- Zhang R, Li Q, Gao J, Qu M, Ding P. 2016. The complete chloroplast genome sequence of the medicinal plant Morinda officinalis (Rubiaceae), an endemic to China. Mitochondrial DNA Part A. 27(6):4324–4325.
- Zhang X-F, Wang J-H, Zhao K-K, Fan W-W, Wang H-X, Zhu Z-X, Wang H-F. 2019. Complete plastome sequence of Hedyotis ovata Thunb. ex Maxim (Rubiaceae): an endemic shrub in Hainan. China. Mitochondrial DNA Part B. 4(1):675–676.