Abstract
In this study, the first complete mitochondrial genome of Myrmeleon formicarius was sequenced. The length of the complete mitochondrial genome of M. formicarius was 16,179 bp with 25.19% GC content. There were 13 protein-coding genes (CDS), 22 transfer RNA genes (tRNA), two ribosomal RNA genes (rRNA), and an AT-rich region. Phylogenetic analysis showed that M. formicarius was closely related to M. immanis. This study provides useful genetic information to better understand the genetic evolution of Myrmeleontidae.
Myrmeleontidae (antlions) belongs to the order Neuroptera, it is widely distributed worldwide and contains 1630 species (Stange Citation2004). However, there are only few studies on complete mitochondrial genome of Myrmeleon immanis sequenced and published to date (Yan et al. Citation2014; Zhang and Wang Citation2016). In this study, the first complete mitochondrial genome of M. formicarius was sequenced and assembled, as well as phylogenies of M. formicarius were constructed by neighbor-joining to understand the evolving relationship.
The M. formicarius larvae were collected from Nanyang, Henan Province, China (32°46′N, 111°31′E) by the traps. The specimens were stored in the Key Laboratory of Integrated Pest Management in Ecological Forests, Fujian Province University, Fujian Agriculture and Forestry University (MF-201909), as well as the total DNA was also stored in this lab. The total DNA was extracted by the TruSeq DNA Sample Preparation Kit (Vanzyme, Nanjing, China) and purified by the QIAquick Gel Extraction Kit (Qiagen GmbH, Germany), then the purified DNA was used with the fragment of ∼300 bp in size for library construction. The mitochondrial genome was sequenced with 150 bp pair-end reads through Illumina Hiseq 2500 by Genesky Biotechnologies Inc. (Shanghai, China). In all, 12,718,748 clean reads were obtained through the quality analysis and filtration by Fastp software (Chen et al. Citation2018). Then, these clean reads were assembled using MtioZ and metaSPAdes software (Nurk et al. Citation2017). The MITOS web server (Matthias et al. Citation2013) was used to annotate the assembly results based on the reference sequence of M. immanis (GenBank accession no. KJ461323). And, tRNA genes were predicted using tRNAscan (Lowe and Eddy Citation1997). The complete mitochondrial genome of M. formicarius was a circular molecule that was 16,179 bp in length (GenBank accession no. MT118664). The GC content of the complete genome was 25.19%, and there were 37 unique genes, including 13 protein-coding genes (CDS), 22 transfer RNA genes (tRNA), two ribosomal RNA genes (rRNA), and an AT-rich region.
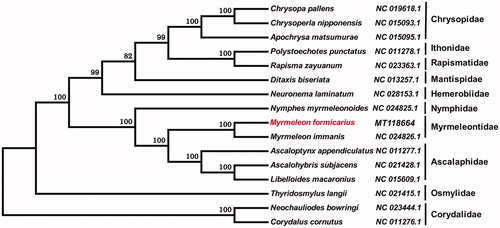
To further investigate the phylogenetic position of M. formicarius, according to the genome sequence of M. formicarius, the phylogenetic analysis was constructed with fifteen different species of Neuropterida by MEGA 6.0 (Tamura et al. Citation2013) using neighbor-joining tree model with 1000 bootstrap replicates. The neighbor-joining tree showed that M. formicarius was closely related to M. immanis, and sister to Ascalaphidae (). The complete mitochondrial genome of M. formicarius will provide useful genetic information to better understand the genetic evolution in Myrmeleon, as well as in Myrmeleontidae and other insects of Neuroptera.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Lowe TM, Eddy SR. 1997. tRNA scan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Matthias B, Alexander D, Frank J, et al. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Stange LA. 2004. A systematic catalog, bibliography and classification of the world antlions (Insecta: Neuroptera: Myrmeleontidae). Mem Am Entomol Institute. 74:1–565.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Yan Y, Wang Y, Liu X, Winterton SL, Yang D. 2014. The first mitochondrial genomes of antlion (Neuroptera: Myrmeleontidae) and split-footed lacewing (Neuroptera: Nymphidae), with phylogenetic implications of Myrmeleontiformia. Int J Biol Sci. 10(8):895–908.
- Zhang J, Wang XL. 2016. The complete mitochondrial genome of Myrmeleon immanis Walker, 1853 (Neuroptera: Myrmeleontidae). Mitochondrial DNA. 27(2):1439–1440.