Abstract
In this study, we present the complete chloroplast genome of Dipterygium glaucum an important medicinal plant in Saudi Arabia, in order to evaluate the evolutionary relationship in the family Cleomaceae. The cp genome is second to be sequenced in Cleomaceae family, it has a circular structure and a length of 158,576 bp with 35.74% GC content. The genome is divided into four regions: LSC of 87,738 bp, SSC of 18,420 bp and a pair of inverted repeats 26,209 bp each. The chloroplast genome of D. glaucum encodes 116 different genes, including 81protien coding genes, 31tRNA genes and four rRNA genes. The phylogenetic relationship showed a close relationship between D. glaucum and Tarenaia hassleriana.
Dipterygium glaucum is a monotypic genus with one species, widely distributed in Saudi Arabia, Egypt, Sudan, and Pakistan (Mehmood et al. Citation2010; Ahmad et al. Citation2014). The species is one of the common traditional plants with multiple medicinal uses. It is popular for the treatment of miss-breathing troubles as trachea dilating agent (Moussa et al. Citation2012), treatment of jaundice, blood purifier, psoriasis, and ringworm infestation and as an antiasthma drug (Rahman et al. Citation2004; Ahmad et al. Citation2014). Shaheen et al. (Citation2017) demonstrate that D. glaucum possesses significant antioxidant, cytotoxic and antimicrobial activities which could be ascribed to its flavonoidal content. Phytochemical studies on D. glaucum revealed the presence of alkaloids, cardiac glycoside, bound anthraquinones, saponins, terpenoids, and sterols (Abdel-Mogib et al. Citation2000; Mehmood et al. Citation2010). The genus Dipterygium has migrated in placement between Brassicaceae (Hutchinson, Citation1967) and Capparaceae (Pax and Hoffmann Citation1936; Hedge et al. Citation1980). Genus has been placed in Capparaceae, subf. Dipterygioideae, based on chemical data the presence of methyl-glucosinolate (Hedge et al. Citation1980; Luning et al. Citation1992). Floral characteristics that ally Dipterygium in Cleomoideae include six stamens of equal length (not tetradynamous) and the presence of a short gynophore.
Chloroplast (cp) DNA plays critical roles in molecular evolution and population genetic studies. We present the assembled sequence of the complete chloroplast cp genome of Dipterygium glaucum (GeneBank accession number: MT041700) for the purpose of providing bases of genetic information for future research.
Fresh leaves of Dipterygium glaucum were collected from Wadi Noa’man mountains, Makkah, Saudi Arabia (21.3502 N 40.1020E). A voucher specimen was prepared and deposited in the herbarium of King Abdulaziz University, Jeddah with the accession number KAU27383. DNA extraction was done using Qiagen DNA extraction kit (Qiagen, Hilden, Germany) based on manufacturer’s guidelines. The Extracted DNA was sequenced using Illumina Hiseq 2500 platform (Novogene Technology, Inc. Beijing, China). Raw data were filtered using PRINSEQ lite Ver0.20.4 to obtained clear reads (5GB). Trimmed sequenced were assembled with NOVOPlasty (Dierckxsens et al. Citation2017). Assembled sequenced were annotated using DOGMA (Wyman et al., Citation2014) which adopt the manual adjustment using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), trNAscan-SE2.0 (Lowe and Chan Citation2016) was used to identify tRNA genes. Finally, the complete chloroplast genome sequence of D. glaucum was submitted to the Genebank with accession number MT041700
The complete plastome genome of D. glaucum is 158,576 bp in length with circular topology; it has 35.74% GC content, Large Single Copy (LSC) 87,738 bp and Small Single copy SSC 18,420 bp. The genome consists of 81 protein coding genes, 31tRNA and four rRNA genes were duplicated within the inverted repeats region. Out of the 116 genes, 14 genes contained one intron and two genes contained two introns.
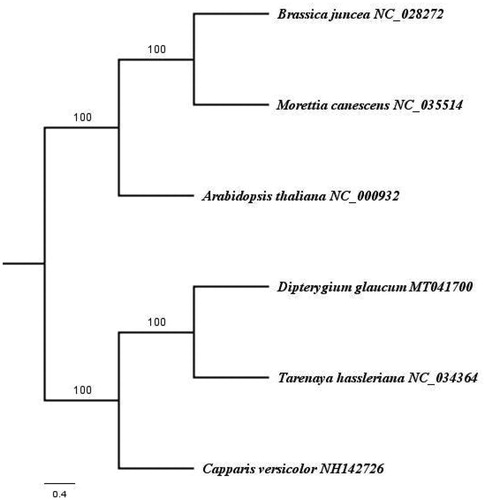
For the purpose of phylogenetic analysis, to evaluate the phylogenetic position of D. glaucum, complete chloroplast genome of 5 species were downloaded from the Genebank: one species from Cleomaceae (Tarenaia hassleriana NC_034364); one species from Capparaceae (Capparis versicolor NH142726) and three species from Brassicaceae (Brassica junces NC_028272, Morettia canescens NC_035514 and Arabidopsis thaliana NV_000932). The downloaded sequences were aligned using MAFFT (Katoh and Standley Citation2013) and the phylogenetic tree was constructed using Maximum parsimony method using PAUP version 4.0b10 (Felsenstein Citation1978) using heuristic searches with 1000 replicates of random taxon addition, tree bisection– reconnection branch swapping, MulTrees on, saving a maximum of 100 trees each replicate. Missing characters were treated as gaps. Support was assessed using 1000 replicates of non-parametric bootstrap analysis.
The result of the phylogenetic analysis showed three distinct clades. The first clade include D. glaucum and T. hassleriana from Cleomaceae, second clade comprising of C. versicolor from Capparaceae and the third clade includes B. junces, M. canescens and A. thaliana from Capparaceae (). The result confirms that placement of D. glaucum in Cleomaceae and is sister to Tarenaia hassleriana.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abdel-Mogib M, Ezmirly ST, Basaif SA. 2000. Phytochemistry of Dipterygium glaucum and Capparis decidua. J Saudi ChemSoc. 4:103–108.
- Ahmad S, Wariss HM, Alam K, Anjum S, Mukhtar M. 2014. Ethnobotanical studies of plantresources of Cholistan desert, Pakistan. Int J Sci Res. 3:1782–1788.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology. 27(4):401–410.
- Hedge IC, Kjaer A, Malver O. 1980. Dipterygium—Cruciferae or Capparaceae? Notes from the Royal Botanical Garden Edinburgh. 38:247–250.
- Hutchinson J. 1967. The genera of flowering plants. Oxford: Clarendon Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lowe T, Chan P. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Luning B, Kers LE, Seffers P. 1992. Methyl glycosinolate confirmed in Puccionia and Dhofaria (Capparidaceae). Biochem. Syst. Ecol. 29:394.
- Mehmood K, Mehmood S, Ramzan M, Arshad M, Yasmeen F. 2010. Biochemical and phytochemical analysis of Dipterygium glaucum collected from Cholistan desert. J Sci Res. 40:13–18.
- Moussa SA, Taia WK, Al-Ghamdy FG. 2012. Acclimation of Dipterygium glaucum Decne. Grown in the Western Coastal part of Saudi Arabia to different water supplies. Int J Res Chem Environ. 2:301–309.
- Pax F, Hoffmann K. 1936. Capparidaceae. In A Engler, K Prantl, editors, Die Natürlichen Pflanzenfamilien, 2nd ed., Vol. 17b. Leipzig: Wilhelm Engelmann, 146–223.
- Rahman MA, Mossa JS, Al-Said MS, Al-Yahya MA. 2004. Medicinal plant diversity in the flora of Saudi Arabia 1: a report on seven plant families. Fitoterapia. 75(2):149–161.
- Shaheen U, Shoeib N, Temraz A, Abdelhady MS. 2017. Flavonoidal constituents, antioxidant, antimicrobial, and cytotoxic activities of Dipterygium glaucum grown in Kingdom of Saudi Arabia. Phcog Mag. 13(51):484.
- Wyman S, Jansen R, Boore J. 2014. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.