Abstract
The family Provannidae is endemic to chemosynthetic environments in the deep-sea. Here, we report the complete mitogenome of a provannid vent snail Alviniconcha boucheti from the North Fiji Basin for the first time. The length of mitogenome was 15,981 bp, with 67.7% AT content. The mitogenome content and its protein-coding gene organization resembled those of other provannid snails. According to the phylogenetic tree, A. boucheti was most closely related to Ifremeria nautilei. Along with this result, further mitogenomic analysis of undetermined taxa within gastropods would improve our understanding in their taxonomic relationship and biogeography.
The abyssochrysoid family Provannidae is endemic to the chemosynthetic environments and composed of 33 extant species belonging to five genera (Chen et al. Citation2019). This family is known to be a paraphyletic taxon based on molecular phylogenetic analysis using seven genetic markers (three mitochondrial and four nuclear) (Johnson et al. Citation2010).
The provannid snail Alviniconcha is characterized by globose and elastic shell decorated with spirally arranged hair (Okutani and Ohta Citation1988). Alviniconcha species are symbiont-hosting snails and usually occur in dense clusters near vent chimneys (Sasaki et al. Citation2010). As of 17 March 2020, the mitochondrial genome (mitogenome) of only two provannid snails, Ifremeria nautilei and Provanna sp., are registered in GenBank. To elucidate the position of Alviniconcha genus in relation to other gastropods in thorough scale, we determined the mitogenome of A. boucheti.
In November 2016, Alviniconcha boucheti samples were collected from the North Fiji Basin (19°03′S and 173°29′E; 2717 m depth) using the ROV ‘ROPOS’ (Canadian Scientific Submersible Facility). Genomic DNA extraction and mitochondrial genome amplification followed the methods of Kim et al. (Citation2018). Library construction and sequencing were performed by Macrogen Service (Macroge, Seoul, Korea) using the Illumina NovaSeq sequencing platform (Illumina, San Diego, CA). The raw data were assembled and refined using NOVOPlasty 2.7.2 (Dierckxsens et al. Citation2017) and Geneious Prime (Biomatters, Auckland, New Zealand). The newly obtained mitochondrial genome was annotated using MITOS (Bernt et al. Citation2013) and adjusted manually. The specimen used for the mitogenome analysis is deposited at the National Marine Biodiversity Institute of Korea (Accession no. MABIK TI00003770).
The complete mitogenome of A. boucheti was 15,981 bp in length (Accession no. MT123331; 67.7% AT content). It contained 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and a non-coding region. The gene organization resembled that of other provannid snails.
All PCGs had an ATN start codon and most of them terminated with a complete stop codon (TAA or TAG) except ND1 and ND5 which had an incomplete stop codon (T––). The length of 16S and 12S rRNAs were 1,395 bp (70.1% AT content) and 973 bp (66.2% AT content), respectively. The length of tRNA genes varied between 65 and 78 bp. A non-coding region (556 bp; 73.0% AT content) was located between the tRNAPhe and COX3.
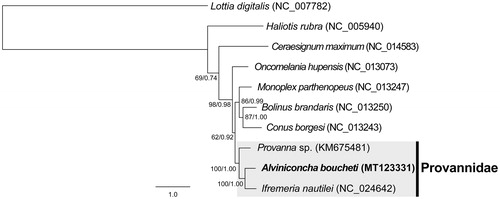
Phylogenetic trees were obtained by maximum likelihood (ML) and Bayesian inference (BI) methods using the 13 PCGs of 10 gastropods (). The tree topology of ML and BI were identical, and overall form was congruent to those from previous studies (Osca et al. Citation2014). According to the tree, the closest relative of A. boucheti was presented to be I. nautilei (100% bootstrap proportions and 1.00 Bayesian posterior probabilities; 74.1% identity for whole mitogenome), which is commonly found with A. boucheti inhabiting the same hydrothermal vents of the southwestern Pacific Ocean.
Figure 1. Phylogenetic relationship of Alviniconcha boucheti and other gastropods based on 13 mitochondrial protein-coding genes using maximum-likelihood (ML) and Bayesian inference (BI) methods. GTR + I + G was selected as the best evolutionary model using jModelTest 2.1.4. Numbers on internodes indicate maximum likelihood bootstrap proportions (left) and Bayesian posterior probabilities (right).
This study contributes in understanding taxonomic relationship and evolution of the deep-sea gastropods. Additional mitogenomic analysis of undetermined taxa in the future would further clarify the relationship within the gastropods.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen C, Kayama Watanabe H, Sasaki T. 2019. Four new deep-sea provannid snails (Gastropoda: Abyssochrysoidea) discovered from hydrocarbon seep and hydrothermal vents in Japan. R Soc Open Sci. 6(7):190393
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Johnson SB, Warén A, Lee RW, Kano Y, Kaim A, Davis A, Strong EE, Vrijenhoek RC. 2010. Rubyspira, new genus and two new species of bone-eating deep-sea snails with ancient habits. Biol Bull. 219(2):166–177.
- Kim S-J, Kang HM, Corbari L, Chan BKK. 2018. First report on the complete mitochondrial genome of the deep-water scalpellid barnacle Arcoscalpellum epeeum (Cirripedia, Thoracica, Scalpellidae). Mitochondrial DNA Part B. 3(2):1288–1289.
- Okutani T, Ohta S. 1988. A new gastropod mollusk associated with hydrothermal vents in the Mariana Back-Arc Basin, Western Pacific. Venus. 47:1–9.
- Osca D, Templado J, Zardoya R. 2014. The mitochondrial genome of Ifremeria nautilei and the phylogenetic position of the enigmatic deep-sea Abyssochrysoidea (Mollusca: Gastropoda). Gene. 547(2):257–266.
- Sasaki T, Warén A, Kano Y, Okutani T, K. Fujikura. 2010. Gastropods from recent hot vents and cold seeps: systematics, diversity and life strategies. In: S. Kiel, ed., The vent and seep biota. Topics in Geobiology, 33:169–254. Dordrecht, Netherlands: Springer.
