Abstract
Juglans cathayensis var. formosana (Hayata) is a precious Juglans tree endemic to China. In this study, we reported its complete chloroplast genome assembled from Illumina pair-end sequencing data. The completed plastome was 159,967 bp in length, including a large-single copy (LSC) region of 89,320 bp, a small-single copy (SSC) region of 18,175 bp, and a pair of inverted repeat (IR) regions of 26,236 bp. The overall GC content of the genome was 36.12%, and the corresponding values of the LSC, SSC, and IR regions were 33.71, 29.79, and 42.43%, respectively. A total of 131 genes were annotated, including 87 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis strongly supported that J. cathayensis var. formosana and J. cathayensis formed a sister clade to J. mandshurica and J. hopeiensis. The newly characterized complete chloroplast genome of J. cathayensis var. formosana will provide essential data for further investigation of this precious species.
Juglans cathayensis var. formosana (Hayata), belonging to the genus Juglans (Juglandaceae), is a precious deciduous broad-leaved tree species endemic to Zhejiang, Jiangsu, Anhui, Jiangxi, Fujian and Taiwan provinces of China. It is considered to be the varietas of J. cathayensis, which can be distinguished by the number of pads on suture. Although complete chloroplast genome sequence of several species from Juglandaceae have previously been unraveled (Hu et al. Citation2016; Dong et al. Citation2017; Hu, Woeste et al. Citation2017; Hu, Yan et al. Citation2017; Yan et al. Citation2017; Ye et al. Citation2018), fundamental genetic information about J. cathayensis var. formosana remains less. In this study, we reported the complete chloroplast genome sequence of J. cathayensis var. formosana based on Illumina pair-end sequencing data, which will provide informative data both for the conservation of this species and for the phylogeny of genus Juglans.
Leaves of J. cathayensis var. formosana were collected from Wuyi Mountains, Fujian Province (27°44′06ʺN, 117°40′13ʺE). The specimen was stored at the State Key Laboratory of Tree Genetics and Breeding (specimen code WysYou-1). The total genomic DNA was extracted using modified CTAB protocol (Wang et al. Citation2015) and sequenced using the Illumina Hiseq X Ten (Illumina, CA, USA). The raw reads were filtered using Trimmomatic (Bolger et al. Citation2014). Clean reads were first aligned to J. cathayensis (NC_033893.1) and then assembled using ABySS v2.1.5 (Jackman et al. Citation2017). The genome was annotated using plann 1.1.2 (Huang and Cronk Citation2015), then adjusted and confirmed with Apollo v2.3.1 (Lee et al. Citation2009). The annotated genomic sequence was then deposited into GenBank with accession number MT012630.
The chloroplast genome of J. cathayensis var. formosana was a quadripartite circular with 159,967 bp, which comprised of a large-single copy (LSC) region of 89,320 bp and a small single copy (SSC) region of 18,175 bp, separated by two inverted repeat (IR) regions of 26,236 bp, respectively. The GC content of the total genome was 36.12%, whereas the IR region had a higher GC content (42.43%) than LSC (33.71%) and SSC (29.79%). The chloroplast genome encoded 131 genes, including 87 protein-coding genes, 36 tRNA genes, and 8 rRNA genes.
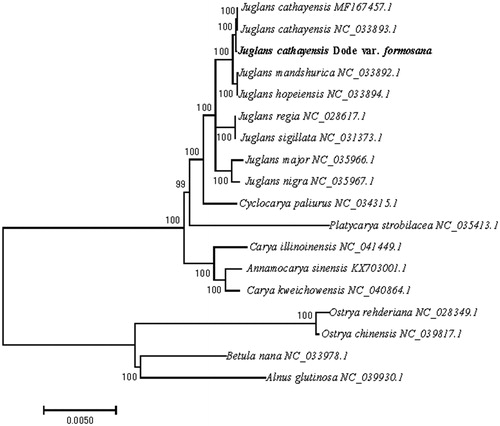
To identify the phylogenetic position of J. cathayensis var. formosana, 13 published chloroplast genomes of Juglandaceae (J. cathayensis, J. mandshurica, J. hopeiensis, J. regia, J. sigillata, J. major, J. nigra, Carya illinoinesis, C. kweichowensis, Annamocarya sinensis, Cyclocarya paliurus and Platycarya strobilacea) and 4 species from Betulaceae (Betula nana, Alnus glutinosa, Ostrya chinensis and O. rehderiana) as outgroups were obtained from NCBI and aligned using MAFFT v7.313 (Katoh and Standley Citation2013). The neighbor-joining (NJ) tree was constructed with RAxML v8.2.11 (Stamatakis Citation2014) and the branch support was estimated with 1000 bootstrap replications (). The results revealed that J. cathayensis var. formosana and J. cathayensis formed a sister clade to J. mandshurica and J. hopeiensis with 100% bootstrap support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics. 30(15):2114–2120.
- Dong W, Xu C, Li W, Xie X, Lu Y, Liu Y, Jin X, Suo Z. 2017. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148.
- Hu Y, Chen X, Feng X, Woeste KE, Zhao P. 2016. Characterization of the complete chloroplast genome of the Endangered species Carya sinensis (Juglandaceae). Conservation Genet Resour. 8(4):467–470.
- Hu Y, Woeste KE, Zhao P. 2017. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front Plant Sci. 7:1955.
- Hu Y, Yan J, Feng X, Dang M, Woeste KE, Zhao P. 2017. Characterization of the complete chloroplast genome of wheel wingnut (Cyclocarya paliurus), an endemic in China. Conservation Genet Resour. 9(2):273–275.
- Huang DI, Cronk Q. 2015. Plann: a command–line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, et al. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27(5):768–777.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee E, Harris N, Gibson M, Chetty R, Lewis S. 2009. Apollo: a community resource for genome annotation editing. Bioinformatics. 25(14):1836–1837.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang H, Pan G, Ma QG, Zhang JP, Pei D. 2015. The genetic diversity and introgression of Juglans regia and J. sigillata in Tibet as revealed by SSR markers. Tree Genet Genomes. 11(1):1
- Yan J, Han K, Zeng S, Zhao P, Woeste K, Li J, Liu ZL. 2017. Characterization of the complete chloroplast genome of Platycarya strobilacea (Juglandaceae). Conservation Genet Resour. 9(1):79–81.
- Ye L, Fu C, Wang Y, Liu J, Gao L. 2018. Characterization of the complete plastid genome of a Chinese endemic species Carya kweichowensis. Mitochondrial DNA B. 3(2):492–493.