Abstract
Rhamnus heterophylla is a small shrub that contained flavonoids, hydroxybenzoic acids, and anthraquinones and flavonoids. It often was used in the treatment of bleeding, irregular menstruation and dysentery. Here, the complete chloroplast genome of the R. heterophylla been constructed from the Illumina sequencing data. The circular cp genome is 156,514 bp in size, and comprises a pair of inverted repeat (IR) regions of 25,276 bp each, a large single-copy (LSC) region of 86,053 bp, and a small single-copy (SSC) region of 19,879 bp. The total GC content is 37.3%, while the corresponding values of the LSC, SSC, and IR regions are 35.3%, 31.7%, and 43.0% respectively. The chloroplast genome contains 130 genes, including 85 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. The neighbor-joining phylogenetic analysis showed a strong sister relationship with Berchemia berchemiifolia and B. wilsonii.
Rhamnus heterophylla is a small shrub that grows on hillside shrubs or forest margins at an altitude of 300–1450 m. It distributes in southeast Gansu, south Shaanxi, Sichuan, Guizhou and Yunnan province in China. Nu-Er-Cha, made by young leaves of R. heterophylla is a well-known herbal tea in China, which contained many active compounds including hydroxybenzoic acids, anthraquinones and flavonoids, and it was used in the treatment of bleeding, irregular menstruation, and dysentery (Wang, Fan, Wang, Wang et al. Citation2019; Wang, Fan, Wang, Yan et al. Citation2019). In spite of its medicinal values, there are few genetic and genomic studies vailable on breeding of this plant. Therefore, we reported the complete chloroplast genome (cp) of R. heterophylla based on Illumina sequencing data, which would be helpful for its evolution and genetics research.
Fresh leaves of R. heterophylla for total genomic DNA extraction were collected in Bashan town, Shaaxi province of China (108°9′19″E, 32°16′48″N). The voucher specimen was preserved at the Herbarium of Xi’an Botanical Garden of Shaanxi Province (accession number XZY19-3098). High-throughput DNA sequencing was conducted on the Illumina HiSeq X ten Sequencing platform (Illumina, CA, USA), and sequenced by Genedenovo (Guangzhou, China). We assembled the cp genome using CLC Genomics Workbench v7.5 (CLC Bio, Aarhus, Denmark). A subset of 42.03 M trimmed reads were used for reconstructing the chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017) with that of its congener Rhamnus taquetii (GenBank: NC_045855) as the initial reference genome. The physical map of the new chloroplast genome was generated using MITObim v1.8 (Hahn et al. Citation2013). Finally, the validated complete chloroplast genome sequence was submitted to the GenBank with accession number MT211599.
The complete chloroplast genome of R. heterophylla is 156,514 bp in size with high coverage (mean 397.2×), containing a pair of inverted repeat (IR) regions of 25,276 bp each, separated by a large single-copy (LSC) region of 86,053 bp and a small single-copy (SSC) region of 19,879 bp. The total GC content is 37.3%, while the corresponding values of the LSC, SSC, and IR regions are 35.3%, 31.7%, and 43.0%, respectively. This chloroplast genome harbors 130 functional genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among them, 44 are involved in photosynthesis and 73 genes are involved in self replication. Seven protein-coding genes, rpl2, rpl23, rps7, rps12, rps19, ndhB, and ycf2, were duplicated genes located in IR regions. Besides, 21 genes contain one intron, such as ndhA, ndhB, petB, and rps12. While accD, chlP, and ycf3 harbor two introns. Summarily, the R. heterophylla cp genome is structurally similar to previously published ones in Rhamnaceae (Choi et al. Citation2015; Jin et al. Citation2020).
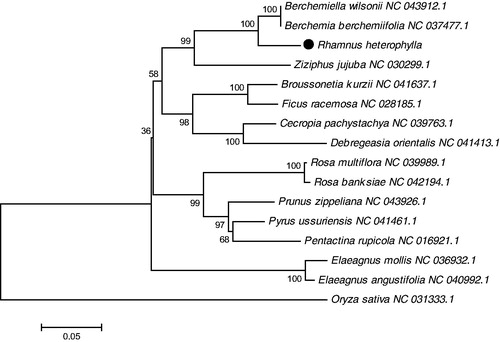
The phylogenetic tree was generated based on the complete cp genome of R. heterophylla and other 14 species in Rhamnaceae (). The alignment was conducted using MEGA 7.0 (Kumar et al. Citation2016). The phylogenetic tree was built using the Neighbor-Joining (NJ) method (Saitou and Nei Citation1987). The results showed that R. heterophylla was closely related to Berchemia berchemiifolia and B. wilsonii. Our findings provide a foundation for further investigation of chloroplast genome evolution in Rhamnaceae.
Figure 1. The phylogenetic tree was constructed using chloroplast genome sequences of 15 species within the Rhamnaceae family and Oryza sativa as an outgroup based on the neighbor-joining method using 500 bootstrap replicates. Chloroplast genome sequences used for this tree are Berchemiella wilsonii NC_043912.1, Berchemia berchemiifolia NC_037477.1, Broussonetia kurzii NC_041637.1, Cecropia pachystachya NC_039763.1, Debregeasia orientalis NC_041413.1, Elaeagnus mollis NC_036932.1, Elaeagnus angustifolia NC_040992.1, Ficus racemosa NC_028185.1, Oryza sativa NC_031333.1, Pentactina rupicola NC_016921.1, Prunus zippeliana NC_043926.1, Pyrus ussuriensis NC_041461.1, Rosa banksiae NC_042194.1, Rosa multiflora NC_039989.1, Ziziphus jujuba NC_030299.1.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Choi KS, Son OG, Park SJ. 2015. The chloroplast genome of Elaeagnus macrophylla and trnH duplication event in Elaeagnaceae. PLOS One. 10(9):e0138727.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data[J]. Nucleic Acids Res. 45(4):e18–e18.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Jin D-P, Park J-W, Park J-S, Choi B-H. 2020. The complete plastid genome of Rhamnus taquetii, an endemic shrub on the Jeju Island of Korea. Mitochondrial DNA Part B. 5(1):924–926.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425.
- Wang L, Fan S, Wang X, Wang X, Yan X, Shan D, Xiao W, Ma J, Wang Y, Li X, et al. 2019. Physicochemical aspects and sensory profiles as various potential factors for comprehensive quality assessment of Nü-Er-Cha produced from Rhamnus heterophylla Oliv. Molecules. 24(18):3211.
- Wang L, Fan SS, Wang XH, Yan X, Xu X, Li X, Wang XP, Li X, Sun SQ, She GM. 2019. Chemical constituents from Rhamnus heterophylla. Chin Tradit Herb Drugs. 50(21):5217–5222.
