Abstract
The seed of Prunus persica is a kind of commonly used Chinese herbal medicine in orthopedics at China. In this study, we assembled and annotated the complete chloroplast genome of P. persica. The chloroplast genome size was 157,704 bp as a circle, which contained a large single-copy region (LSC, 85,883 bp), a small single-copy region (SSC, 19,061 bp) and a pair of inverted repeat regions (IR, 26,380 bp). The overall nucleotide composition is: 31.1% of A (49,092 bp), 32.1% T (50,709 bp), 18.8% C (29,625 bp) and 18.0% G (28,278 bp), with a total G + C content of the chloroplast genome 36.8% and A + T of 63.2%. The chloroplast genome of P. persica contains 123 genes, which included 78 protein-coding genes (PCG), 37 transfer RNA (tRNAs) and 8 ribosome RNA (rRNAs). Furthermore, the phylogenetic analysis based on 14 plants species confirmed the position of P. persica closely related to Prunus mongolica.
Prunus persica belongs to the family Rosaceae. The seeds of Prunus persica (Tao-Ren in Chinese) are well known as a Chinese herbal medicine in China and other Asian countries (Rho et al. Citation2008). The seed of P. persica contains amygdalin, prunasin, tryptophane, glucose, sucrose, chlorogenic acid, triolein and so on. Besides, peach seed oil is rich in unsaturated fatty acids, mainly including oleic acid and linoleic acid. The seeds of P. persica have a strong anti-inflammatory and analgesic pharmacological activity (Ester et al. Citation2018). Now, less information about the genome of the seed of Prunus persica was published. They are frequently used as an ingredient in a variety of Chinese herbal medicine prescriptions, particularly those used to treat orthopedics disease. In this study, we assembled and annotated the complete chloroplast genome of P. persica and discussed phylogenetic relationship with other plants. It will be useful for study the medicinal valuable and research the drug development the seed of P. persica in future.
The fresh sample of Prunus persica was purchased in Shimane University School of Medicine (127.80E; 26.34 N). The seeds of P. persica and total genomic DNA were extracted using Plant Tissues Genomic DNA Extraction Kit (TaKaRa, DL, and CN) and stored in Shimane University School of Medicine (No. SUSM01). The cp genomic DNA of P. persica was purified and fragmented using the NEB Next UltraTM II DNA Library Prep Kit (NEB, BJ, CN) that the cp genomic was sequenced. Quality reads and adapters control was performed and removed low-quality reads and adapters using the NGS QC Toolkit software (Patel and Jain Citation2012). The chloroplast genome was assembled and annotated using the MitoZ software (Meng et al. Citation2019). The physical map of the new chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013).
The chloroplast (cp) genome of Prunus persica was 157,704 bp in size as a circle (NK1487411), which contained a large single-copy region (LSC) of 85,883 bp, a small single- copy region (SSC) of 19,061 bp and a pair of inverted repeat regions (IRA and IRB) of 26,380 bp. The cp genome of P. persica comprised 123 genes, which included 78 protein-coding genes (PCG), 37 transfer RNA genes (tRNA) and 8 ribosomal RNA genes (rRNA). In the IR regions, 19 genes were found duplicated, including 8 PCG species (rps19, rpl2, rpl23, ycf2, ndhB, rps7, rps12 and ycf1), 7 tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRNA species (rrn16, rrn23, rrn4.5 and rrn5). The overall nucleotide composition is: 31.1% of A (49,092 bp), 32.1% T (50,709 bp), 18.8% C (29,625 bp) and 18.0% G (28,278 bp), that is the total G + C content of 36.8% and A + T of 63.2%.
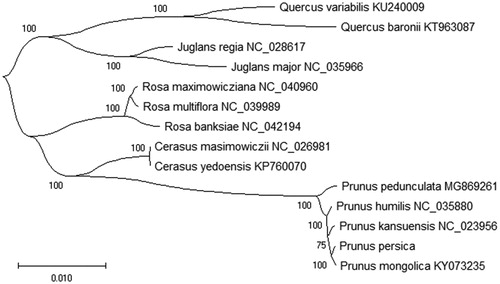
Phylogenetic analysis using the Maximum-Likelihood (ML) methods, we analyzed the relationship of other 13 plant species chloroplast genomes with Prunus persica. The phylogenetic tree was reconstructed using ML methods and was performed using RAxML software (Stamatakis Citation2014) with the GTR + G + I model. Branch support was inferred using 2,000 bootstrap replicates. The phylogenetic tree was represented using MEGA X (Kumar et al. Citation2018) and edited using FigTree version 1.4.4. Furthermore, the phylogenetic ML tree result indicated that the chloroplast genome of Prunus persica is clustered and closest to Prunus mongolica in the evolutionary relationship (). This study can be useful for study the medicinal valuable and research the drug development the seed of P. persica in future.
Figure 1. The phylogenetic tree based on the 14 plant species complete chloroplast genome sequences using the Maximum-Likelihood (ML) method (bootstrap repeat is 2,000). The length of branch represents the divergence distance. All the plant species chloroplast genomes in this study have been deposited in the GenBank.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ester HC, Marina ML, Ma CG. 2018. Multiple protective effect of peptides released from Olea europaea and Prunus persica seeds against oxidative damage and cancer cell proliferation. Food Res Int. 106:458–467.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evolut. 35(6):1547–1549.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Meng GL, Li YY, Yang CT, Liu SL, 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Rho J-R, Jun C-S, Ha Y-A, Yoo M-J, Cui M-X, Baek H-S, Lim J-A, Lee Y-H, Chai K-Y. 2008. Cheminform abstract: isolation and characterization of new alkaloid from the seed of Prunus persica L. and its antiinflammatory activity. ChemInform. 39(2):1289–1293.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
