Abstract
The complete plastome sequence of Asplenium × bimixtum (Aspleniaceae) was determined. The whole genome is 153,098 bp in length, consisting of large (86,394 bp) and small (21,508 bp) single-copy regions, and a pair of identical inverted repeats (22,598 bp). The genome contains 88 protein-coding genes, 33 tRNA genes, 8 rRNA genes, and 1 pseudogene (infA). The phylogenetic analysis based on 84 protein-coding genes showed that Asplenium is monophyletic with high bootstrap value.
The spleenwort genus Asplenium L. (Aspleniaceae) is one of the largest genera of ferns comprising more than 700 species and distributed nearly all over the world (PPG I Citation2016). Reticulate evolution of Asplenium species is well known and many cases are thoroughly investigated and documented, such as Appalachian Asplenium hybrid complex (Wagner Citation1954; Smith and Levin Citation1963) and New Zealand Asplenium hybrids (Brownsey Citation1977; Perrie and Brownsey Citation2005; Shepherd et al. Citation2008). In the Korean peninsula, four purported Asplenium hybrids have been recognized and A. ruprechtii Sa. Kurata has been presumed to be one of the parental species of all of those hybrids (Lee and Lee Citation2015; Korea National Arboretum Citation2017). Especially, A. ×bimixtum C. S. Lee & K. Lee ex Y.-H. Ha, recently validated hybrid name from nomenclatural issues, was suggested as a hybrid derived from three Asplenium species (i.e. A. ruprechtii, A. incisum, and A. trichomanes; Lee et al. Citation2015; Ha et al. Citation2020). However, more thorough and cautious investigations are needed to elucidate the reticulate evolution of Korean Asplenium complex. Therefore, characterization of chloroplast genome sequence of A. ×bimixtum will contribute to identify maternal parent lineage of this hybrid.
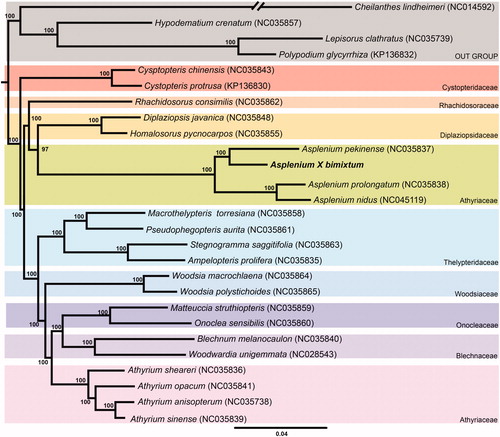
Wild individual was collected from Yeoncheon-Gun, Gyeonggi-do, Republic of Korea (Voucher specimen Hyh149 deposited in the Herbarium of Korea National Arboretum, KH). A total genomic DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA). We constructed library and sequenced using the Illumina Miseq platform (Illumina, San Diego, California, USA). Produced 7,695,796 raw reads were trimmed (0.01% error probability per base) using the Geneious v. 10.2.2 program (Biomatters Ltd., Auckland, New Zealand). Reference guide assembly was performed and redundancy was checked with the chloroplast genome of A. pekinense (NC_035837). Genome annotation of the draft sequence was verified with Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). RNA editing by C-to-U conversions (Knie et al. Citation2016) were annotated using the Geneious R v.10.2.2 program. Maximum likelihood (ML) analysis was performed using concatenated 84 protein-coding genes of 27 species (23 species of Eupolypod II Ferns and 4 species of Eupolypod I). The phylogenetic tree was constructed by IQTREE v.1.6.8 (Nguyen et al. Citation2015) with 1000 bootstrap replications. The complete chloroplast genome of A. ×bimixtum (Genbank accession is MN912732) was 153,098 bp in length. The mean coverage of trimmed reads aligned to genome sequence with strict option (Maximum mismatches per read: 0%, Maximum ambiguity: 1) in Geneious program was 294×. The plastome has four subregions: large (86,394 bp) and small (21,508 bp) single-copy regions and a pair of identical inverted repeat (IR) regions (22,598 bp). It contains 130 genes consisting of 88 protein-coding genes, 33 tRNA genes, 8 rRNA genes, and 1 pseudogene (infA). Duplicated genes in IR were with four protein-coding genes, six tRNA genes, and four rRNA genes. The ML tree showed that the family Aspleniaceae is monophyletic (100% BS, ) and the sister relationship between A. ×bimixtum and A. pekinense is strongly supported (100% BS, ).
Figure 1. Maximum likelihood tree inferred from combined 84 protein-coding genes of 27 Eupolypod plastomes including four species of Eupolypod I (Cheilanthes lindheimeri, Hypodematium crenatum, Lepisorus clathratus, Polypodium glycyrrhiza) as outgroups. The bootstrap support values are shown at the branches.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Brownsey PJ. 1977. Asplenium hybrids in the New Zealand flora. NZ J Bot. 15(3):601–637.
- Ha Y-H, Son DC, Kim D-K. 2020. Validation of a fern hybrid from Korea, Asplenium× bimixtum (Aspleniaceae). Phytotaxa. 433(4):295–296.
- Knie N, Grewe F, Fischer S, Knoop V. 2016. Reverse U-to-C editing exceeds C-to-U RNA editing in some ferns – a monilophyte-wide comparison of chloroplast and mitochondrial RNA editing suggests independent evolution of the two processes in both organelles. BMC Evol Biol. 16(1):134.
- Korea National Arboretum. 2017. Checklist of vascular plants in Korea. Pocheon: Korea National Arboretum.
- Lee CS, Lee KH. 2015. Pteridophytes of Korea: Lycophytes & ferns. Seoul: Geobook.
- Lee CS, Lee K, Yeau SH, Chung K-S. 2015. Two new and one unrecorded natural hybrids between Asplenium ruprechtii and related taxa (Aspleniaceae). Korean J Pl Taxon. 45(4):362–368.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Perrie LR, Brownsey PJ. 2005. Insights into the biogeography and polyploid evolution of New Zealand Asplenium from chloroplast DNA sequence data. Am Fern J. 95(1):1–21.
- Ppg I. 2016. A community‐derived classification for extant lycophytes and ferns. J Syst E. 54:563–603.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server):W686–W689.
- Shepherd LD, Perrie LR, Brownsey PJ. 2008. Low-copy nuclear DNA sequences reveal a predominance of allopolyploids in a New Zealand Asplenium fern complex. Mol Phylogenet Evol. 49(1):240–248.
- Smith DM, Levin DA. 1963. A chromatographic study of reticulate evolution in the Appalachian Asplenium complex. Am J Bot. 50(9):952–958.
- Wagner WH. 1954. Reticulate evolution in the Appalachian Aspleniums. Evolution. 8(2):103–118.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
