Abstract
The complete mitochondrial genome was sequenced from the NIES strain of the freshwater water flea Daphnia magna. The sequenced mitochondrial genome size of the NIES strain of D. magna was 17,432 bp, possessing identical gene order and the contents to those of the congeneric species Daphnia galeata in the genus Daphnia, but 16S and 12S rRNA genes was in opposite direction compared to other daphnid, while ND2 gene was also often found in opposite direction in the genus Daphnia. Overall, the mitochondrial genome of D. magna NIES strain has 13 protein-coding genes (PGCs), two rRNAs, and 22 tRNAs. Of 13 PGCs, four genes (CO1, CO2, ND5, and ND4) had incomplete stop codons. Furthermore, the stop codons of the remaining nine PGCs were TAG and TAA, while the start codons of CO1, ND6, and ND3 genes were ATT, ATT, and ATC, respectively. The base composition of D. magna NIES strain mitogenome shows an anti-G and –T bias (16.5% and 16.0%) on the L strand, respectively.
Of more than 200 daphnid species (Kotov et al. Citation2012), the complete mitochondrial genome of the freshwater water flea Daphnia pulex has been firstly reported (Crease Citation1999) but had the high mutation rates in mitochondrial genome (Xu et al. Citation2012). The freshwater water flea Daphnia magna is one of the promising model organisms in the freshwater environment and has been widely used in genetics (Kumagai et al. Citation2017), genomics (Lee et al. Citation2019), developmental biology (Ismail et al. Citation2018; Kim et al. Citation2018), environmental toxicology (Oda et al. Citation2006), and in other applied aspects. In this paper, we report the complete mitochondrial genome of the freshwater water flea D. magna NIES strain as a life barcode that can be useful to better understand the phylogenetic position of and within the genus Daphnia.
The water flea D. magna (NIES strain) was originated from the United States Environmental Protection Agency’s Environmental Research Laboratory in Duluth, Minnesota via the Chemicals Evaluation and Research Institute, Japan (Oda et al. Citation2006) and maintained for about 34 years at the National Institute for Environmental Studies (NIES) in Tsukuba and other institutions in Japan. We obtained D. magna (NIES strain) from Osaka University in Japan (kindly provided by Prof. Hajime Watanabe) on July 31, 2019 and maintained in the Department of Biological Sciences, Sungkyunkwan University in South Korea. De novo assembly was initially performed with Nanopore sequences using SMARTdenovo (https://github.com/ruanjue/smartdenovo). Nanopore sequences data was again mapped on the initial de novo assembly using Medaka (https://github.com/nanoporetech/medaka) to correct errors in nanopore sequences and create a consensus nanopore sequences. Illumina reads including 500 bp paired end sequencing were mapped to nanopore assembly to correct and polish the consensus sequences using Pilon (Walker et al. Citation2014) (https://github.com/broadinstitute/pilon/wiki). Of the assembled 618 D. magna NIES strain contigs, a single contig was mapped to the mitochondrial DNA of D. magna KIT (GenBank accession no. MK370029).
The total length of the complete mitochondrial genome of D. magna NIES strain is 17,432 bp (GenBank accession no. MT199637). The mitochondrial genome of D. magna NIES strain contains 13 protein-coding genes (PGCs), two rRNAs, and 22 tRNAs. The direction of 13 PGCs of D. magna NIES strain was identical to those of a congeneric species Daphnia galeata (Tokishita et al. Citation2017) but the direction of two rRNA genes was opposite. Of 13 PCGs in D. magna NIES strain, four genes (CO1, CO2, ND5, and ND4) had incomplete stop codons. The mitochondrial genome base composition of 13 PCGs was 26.60% for A, 43.00% for T, 12.75% for G and 17.65% for C. The A + T base composition (69.6%) was slightly higher than G + C base composition (30.4%). Particularly, in D. magna NIES strain, there is an anti-T (17.7% and 16.0%) and anti-G bias (18.2% and 16.5%) at the second and third position of codons on the L strand. The most commonly found start codon was ATG but some genes used ATT (CO1 and ND6 genes) and ATC (ND3 gene), respectively in D. magna NIES strain.
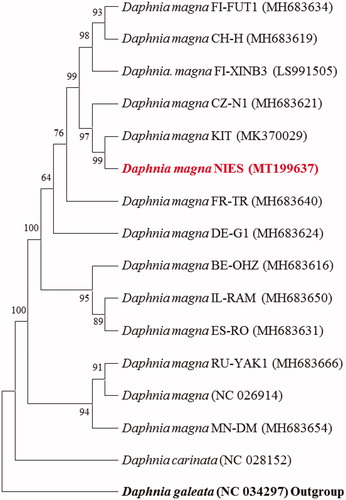
Of 13 D. magna strains with known complete mitogenomes, the placement of D. magna NIES strain with an outgroup (D. galeata) was shown in . In 16 daphnids, nearly all components (12 PGCs, 22 tRNAs) were identically placed but the direction of ND2 gene of four strains (xinb3, KIT, NIES, and GenBank no. NC_026914) of D. magna was in forward direction as shown in other two congeneric species (D. carinata and D. galeata) (Jeong et al. Citation2019). In addition, the direction of two rRNAs was opposite compared to other daphnids.
Figure 1. Phylogenetic analysis. We conducted a comparison of the mitochondrial genomes of 16 daphnids. Twelve protein-coding genes of each mitochondrial genome except for ND2 gene were aligned using MEGA software (ver. 7.0) with the ClustalW alignment algorithm. The best-fit substitution model for phylogenetic analysis was determined with the lowest Bayesian Information Criterion and Akaike Information Criterion scores using a maximum likelihood (ML) analysis. Maximum likelihood phylogenetic analyses were conducted using RAxML (ver. 8.2.8) with the LG + G + I model. The bootstrap analysis was conducted with 1000 replications. Ln = −16,797.57. Modified from Jeong et al. (Citation2019) Mito. DNA B 4:1021-1022.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Crease TJ. 1999. The complete sequence of the mitochondrial genome of Daphnia pulex (Cladocera: Crustacea). Gene. 233(1–2):89–99.
- Ismail NIB, Kato Y, Matsuura T, Watanabe H. 2018. Generation of white-eyed Daphnia magna mutants lacking scarlet function. PLoS One. 13(11):e0205609.
- Jeong C-B, Choi B-S, Hwang D-S, Choi J, Lee J-S. 2019. Complete mitochondrial genome of the water flea Daphnia magna (Cladocera, Daphniidae). Mitochondrial DNA B. 4(1):1021–1022.
- Kim D-H, Lee B-Y, Kim H-S, Jeong C-B, Hwang D-S, Kim I-C, Lee J-S. 2018. Identification and characterization of homeobox (Hox) genes and conservation of the single Hox cluster (324.6 kb) in the water flea Daphnia magna. J Exp Zool (Mol Dev Evol). 330(2):76–82.
- Kotov A, Forró L, Korovchinsky NM, Petrusek A. 2012. Crustacea-Cladocera checklist. World checklist of freshwater Cladocera species. Belgian Biodiversity Platform [cited 2012 Oct 29]. Available from: http://fada.biodiversity.be/group/show/17
- Kumagai H, Nakanishi T, Matsuura T, Kato Y, Watanabe H. 2017. CRISPR/Cas-mediated knock-in via non-homologous end-joining in the crustacean Daphnia magna. PLoS One. 12(10):e0186112.
- Lee B-Y, Choi B-S, Kim M-S, Park JC, Jeong C-B, Han J, Lee J-S. 2019. The genome of the freshwater water flea Daphnia magna: a potential use in molecular ecotoxicology. Aquat Toxicol. 210:69–84.
- Oda S, Tatarazako N, Watanabe H, Morita M, Iguchi T. 2006. Genetic differences in the production of male neonates in Daphnia magna exposed to juvenile hormone analogs. Chemosphere. 63(9):1477–1484.
- Tokishita SI, Shibuya H, Kobayashi T, Sakamoto M, Ha JY, Yokobori SI, Yamagata H, Hanazato T. 2017. Diversification of mitochondrial genome of Daphnia galeata (Cladocera, Crustacea): Comparison with phylogenetic consideration of the complete sequences of clones isolated from five lakes in Japan. Gene. 611:38–46.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Xu S, Schaack S, Seyfert A, Choi E, Lynch M, Cristescu ME. 2012. High mutation rates in the mitochondrial genomes of Daphnia pulex. Mol Biol Evol. 29(2):763–769.
