Abstract
Acer triflorum (Sapindaceae: Acer) is a deciduous arbor species with excellent ornamental values because of its reddish-tan exfoliating bark and attractive three leaflets. However, due to some wild populations being threatened by poor seed viability, it was listed as vulunerable in China. Its complete chloroplast (cp) genome was obtained using genome Illumina pair-end sequencing data. The cp genome was 156,024 bp in length with a typical circular structure, containing a large single copy (LSC) region of 85,815 bp and a small single copy (SSC) region of 18,051 bp, which were separated by a pair of inverted repeats (IRs) (26,079 bp). A total of 136 genes were annotated, including 88 protein-coding genes, 40 tRNAs and 8 rRNAs. The overall GC content was 37.9%. The phylogenetic analysis suggested that A. triflorum was the most closely related to A. griseum. The complete chloroplast genome of A. triflorum is useful for assessment of genetic diversity and further for taxonomic study of Acer L.
Acer triflorum is a deciduous arbor species, which belongs to the section Trifoliata in the genus Acer of Sapindaceae, mainly distributed in northeastern China and Korea. It is well known and valued not only for its reddish-tan exfoliating bark, but also for its attractive three leaflets, which may turn green to brilliant orange, purple, gold or scarlet in autumn (Twombly Citation2004). Previous scholars have conducted some studies on the constituents of stem bark (Nagai et al. Citation1990) and the development of SSR primers (Wang et al. Citation2018). However, some wild populations of A. triflorum are threatened by poor seed viability, thus it was listed as vulunerable in China (Gibbs and Chen Citation2009). Therefore, the complete cp genome of A. triflorum was assembled and surveyed in order to understanding its phylogenetic position.
Fresh leaves of A. triflorum were collected from a wild individual tree from Tonghua County, Jilin Province, China (N41°44′49′′, E125°59′13′′). The DNA specimen was deposited in Laboratory of Forest Silviculture and Tree Cultivation, Research Institute of Forestry, Chinese Academy of Forestry, China (Voucher specimen: ACTRI-JITH2019-01). The total genomic DNA was extracted using a plant genomic DNA extraction kit (DP350) (Tiangen biotech Inc., Beijing, China) and sequenced using the Illumina Hiseq Platform (Huitong biotechnology Inc., Shenzhen, China). The de novo assembly and annotation of the chloroplast genome were performed using SPAdes v3.9.0 and DOGMA. A chloroplast genome map was drawn using OGDRAW.
Similar to most higher plants, the complete chloroplast genome of A. triflorum had a typical quadripartite structure with 156,024 bp long, containing a large single copy (LSC) region of 85,815 bp and a small single copy (SSC) region of 18,051 bp, which were separated by a pair of inverted repeats (IRs) (26,079 bp). The overall GC content was 37.9%. As a result of annotation,136 genes were identified, including 40 tRNAs, 8 rRNAs and 88 protein-coding genes.
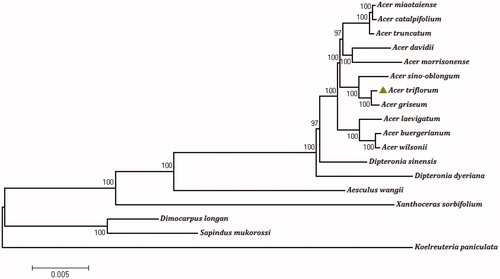
The phylogenetic relationships was reconstructed based on the complete chloroplast genomes of 18 species, using maximum-likelihood (ML) analysis based on GTR + I model in MEGA v 7.0.14 (Kumar et al. Citation2016). All sequences were aligned using MAFFT (Nakamura et al. Citation2018). The result indicated that A. triflorum was the most closely phylogenetic related to A. griseum and they were within the same clade, which is different to the other congeneric taxa from the other sections. This is in accord with morphological taxonomy, which they are relative species and both in the section Trifoliata. Moreover, A. triflorum and A. griseum were clusted with other 9 Acer taxon as a monophyly by 100% bootstrap values and both were sisters to the remaining Acer taxa, consistent with the result of Wang et al. (Citation2017). However, the phylogenetic analysis result based on 11 Acer species () did not support A. griseum as the basal-most species in Acer lineage (Wang et al. Citation2017). The complete chloroplast genome of A. triflorum is useful for assessment of genetic diversity and further for taxonomic study of Acer L.
Figure 1. Phylogenetic tree reconstruction of 18 species using maximum likelihood (ML) based on the complete chloroplast genome sequences of A. triflorum and other 17 species. There are the bootstrap support values from 1000 replicates given at each node. Their accession numbers are as follows: Acer buergerianum: NC_034744; Acer catalpifolium: NC_041080; Acer davidii: NC_030331; Acer griseum: NC_034346; Acer laevigatum: NC_042443; Acer miaotaiense: NC_030343; Acer morrisonense: NC_029371; Acer sino-oblongum: NC_040106; Acer truncatum: NC_037211; Acer wilsonii: NC_040988; Aesculus wangii: NC_035955; Dimocarpus longan: NC_037447; Dipteronia dyeriana: NC_031899; Dipteronia sinensis: NC_029338; Koelreuteria paniculata: NC_037176; Sapindus mukorossi: NC_025554; Xanthoceras sorbifolium: NC_037448.
Data availability
The complete chloroplast genome sequence of A. triflorum was submitted to Genebank of NCBI with the accession number MN602455.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Gibbs D, Chen YS. 2009. The red list of maples. Richmond (UK): Botanic Gardens Conservation International.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Nagai M, Matsuda E, Inoue T, Fujita M, Chi HJ, Ando T. 1990. Studies on the Constituents of aceraceae plants. VII.: diarylheptanoids from Acer griseum and Acer triflorum. Chem Pharm Bull. 38(6):1506–1508.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Twombly K. 2004. Acer triflorum. Horticulture. 101:A4.
- Wang HY, Wei BY, Xiao HX. 2018. Development of 15 microsatellite markers in Acer triflorum (Aceraceae) and cross-amplification in congeneric species. Appl Plant Sci. 6(7):e01166.
- Wang WC, Chen SY, Zhang XZ. 2017. The complete chloroplast genome of the endangered Chinese paperbark maple, Acer griseum (Sapindaceae). Conserv Genet Resour. 9(4):527–529.
