Abstract
Chicoreus asianus is an ecologically and economically important species of Muricidae, which comprises a highly diverse group of carnivorous marine snails. However, the taxonomic classification and phylogenetic studies have so far been limited. In this study, we report the complete mitochondrial genome of C. asianus. The mitogenome has 15,361 base pairs (65.2% A + T content) and made up of total of 37 genes (13 protein-coding, 22 transfer RNAs and 2 ribosomal RNAs), and a control region. The complete mitogenomes of C. asianus. will provide useful genetic information for future taxonomic and phylogenetic classification of Neogastropoda.
With over 1600 described species and inhabited worldwide from tropical to polar seas, the Muricidae is one of the most important families in the Neogastropoda (Zou et al. Citation2011). As carnivorous marine snails, muricids play an important role in structuring marine benthic ecosystem (Barco et al. Citation2010). Moreover, some muricids including Chicoreus asianus are economically important as delicious seafood and traditional medicinal materials (Ramasamy et al. Citation2013; Zhong et al. Citation2019a). However, the phylogeny and classification of Muricidae are still debated due to the morphological convergence and plasticity (Barco et al. Citation2010; Zou et al. Citation2011). The complete mitochondrial genome is an enhance resolution to recover phylogenetic relationships in Muricidae, but mitogenome information in Muricidae is still inadequate (Zhong et al. Citation2019b). Here, we report the complete mitochondrial genome sequence of C. asianus, which will provide a better insight into phylogenetic assessment in Muricidae.
A tissue samples of C. asianus from five individuals were collected from GuangXi province, China (Beihai, 21.45098 N, 109.521139 E), and the whole body specimen (#GR0172) were deposited at Marine biological Herbarium, Guangxi Institute of Oceanology, Beihai, China. The total genomic DNA was extracted from the muscle of the specimens using an SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. DNA libraries (350 bp insert) were constructed with the TruSeq NanoTM kit (Illumina, San Diego, CA) and were sequenced (2 × 150 bp paired-end) using HiSeq platform at Novogene Company, China. Mitogenome assembly was performed by MITObim (Hahn et al. Citation2013). The complete mitogenome of Chicoreus torrefactus (GenBank accession number: NC_039164) was chosen as the initial reference sequence for MITObim assembly. Gene annotation was performed by MITOS (http://mitos2.bioinf.uni-leipzig.de/).
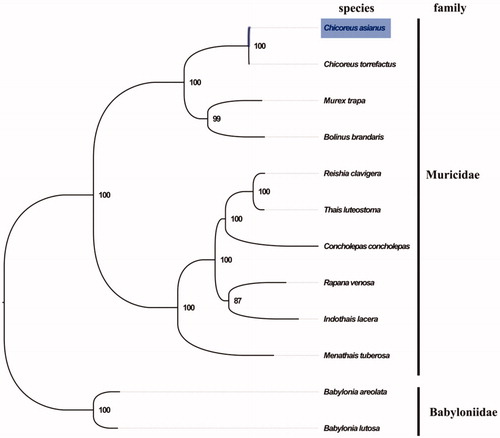
The complete mitogenome of C. asianus was 15,361 bp in length (GenBank accession number: MN793976), and containing the typical set of 13 protein-coding, 22 tRNA and 2 rRNA genes, and a putative control region. The overall base composition of the mitogenome was estimated to be A 27.9%, T 37.6%, C 16.0% and G 18.5%, with a high A + T content of 65.2%, which is similar, but slightly lower than Murex trapa (66.3%) (Zhong et al. Citation2019b). The mitogenomic phylogenetic analyses showed that C. asianus and C. torrefactus clustered in a single clade then clustered with M. trapa with high bootstrap value (), which is consistent with the phylogenetic analyses of Muricidae using the entire 18S rRNA, histone H3, and three partial mitochondrial genes (Zou et al. Citation2011). Our result was consistent with the taxonomic classification of Muricidae, which will contribute to further phylogenetic and comparative mitogenome studies of Muricidae, and related families.
Figure 1. Phylogenetic tree of 12 species in order Neogastropoda. The complete mitogenomes is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 100 bootstrap replicates. The bootstrap values were labeled at each branch nodes. The gene's accession number for tree construction is listed as follows: Babylonia areolata (NC_023080), Babylonia lutosa (NC_028628), Chicoreus torrefactus (NC_039164), Murex trapa (MN462589), Bolinus brandaris (NC_013250), Menathais tuberosa (NC_031405), Concholepas concholepas (NC_017886), Thais luteostoma (NC_039165), Reishia clavigera (NC_010090), Rapana venosa (NC_011193), and Indothais lacera (NC_037221).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Barco A, Claremont M, Reid DG, Houart R, Bouchet P, Williams ST, Cruaud C, Couloux A, Oliverio M. 2010. A molecular phylogenetic framework for the Muricidae, a diverse family of carnivorous gastropods. Mol Phylogenet Evol. 56(3):1025–1039.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Ramasamy P, Thampi DPK, Chelladurai G, Gautham N, Mohanraj S, Mohanraj J. 2013. Screening of antibacterial drugs from marine gastropod Chicoreus ramosus. J Coastal Life Med. 1:181–185.
- Zhong SP, Huang GQ, Liu YH, Huang LH. 2019a. The complete mitochondrial genome of marine gastropod Melo melo (neogastropoda: volutoidea). Mitochondrial DNA Part B. 4(2):4161–4162.
- Zhong SP, Huang LH, Huang GQ, Liu YH, Wang WX. 2019b. The first complete mitochondrial genome of Murex from Murex trapa (Neogastropoda: Muricidae). Mitochondrial DNA Part B. 4(2):3394–3395.
- Zou S, Li Q, Kong L. 2011. Additional gene data and increased sampling give new insights into the phylogenetic relationships of Neogastropoda, within the caenogastropod phylogenetic framework. Mol Phylogenet Evol. 61(2):425–435.
