Abstract
Plantago media, known as the hoary plantain, is a species of flowering plant in the plantain family Plantaginaceae that also was used as one of the Chinese herb in China. In this study, P. media chloroplast genome sequence was 164,130 base pairs (bp) as a circular that had a typical quadripartite structure. It contained a large single-copy region (LSC) of 82,758 bp, a small single-copy region (SSC) of 4,578 bp, and a pair of inverted-repeat (IR) regions of 38,397 bp. The overall nucleotide composition of chloroplast genome is: A (30.7%), T (31.3%), C (19.2%), G (18.8%), and the total G + C content of 38.0%. The total 140 genes were found in this chloroplast genome sequence that included 94 protein-coding genes (PCG), 38 transfer RNA (tRNA) genes, and 8 ribosome RNA (rRNA) genes. In phylogenetic analysis, the result showed that P. media is closely related to P. depressa of the family Plantaginaceae using the maximum-likelihood (ML) method in the phylogenetic relationship.
Plantago media belongs to the genus Plantago and the family Plantaginaceae, which is an economically and medicinally important species, least is known about its genomics data and evolution information. Plantago media is used in traditional medicine around the world, which is astringent, anti-toxic, antimicrobial, anti-inflammatory, anti-histamine, as well as a demulcent, expectorant, styptic, and diuretic (Asaf et al. Citation2020). Plantago media contains mucilage, tannins, and iridoid glycosides, which is believed to give it antimicrobial, anti-inflammatory, and expectorant properties. Apart from having medicinal benefits, the leaves are nutritious too. They contain iron, calcium, potassium, and vitamins A, K, C and B. Add a few young tender leaves to a fresh salad or cook them like you would a leafy green to benefit from this nourishing plant (Sams 2015). Plantago media is very important and beneficial for people in the treatment and prevention of some diseases, fewer genomics data and evolution information of P. media about genome data and study on the evolution of the family Plantaginaceae species. In this study, we obtained the chloroplast genome of Plantago media that can help to research the relationship between origin and evolution, also can be useful valuable molecular dates of Chinese herb for further research.
In this study, the Plant Tissues Genomic DNA Extraction Kit (TIANGEN, BJ and CN) was used to isolate the chloroplast genome DNA from the fresh P. media that was collected from the market of herb near the Third Hospital of Nanchang and located at Nanchang, Jiangxi, China (28.66 N, 115.90E). The chloroplast genome DNA of P. media was stored in the Third Hospital of Nanchang (No. THNC-01). The chloroplast genome DNA was purified and sequenced, which was quality controlled and removed to the collected raw sequences by FastQC (Andrews Citation2015). The chloroplast genome of P. media was assembled by NOVOPlasty (Dierckxsens et al. Citation2017) and annotated by Geneious 8.1.7 (Kearse et al. Citation2012). The genes in chloroplast genome were predicted using CPGAVAS (Liu et al. Citation2012) and corrected by NCBI Blast search. The chloroplast genome sequence of P. media was submitted into NCBI database with the GenBank accession number of NK8904461.
Plantago media chloroplast genome sequence was 164,130 base pairs (bp) in size that had a typical quadripartite structure. It contained a large single-copy region (LSC) of 82,758 bp, a small single-copy region (SSC) of 4578 bp and a pair of inverted repeat (IR) regions of 38,397 bp. The overall nucleotide composition of chloroplast genome sequence is: A (30.7%), T (31.3%), C (19.2%), G (18.8%) and the total G + C content of 38.0%. Plantago media chloroplast genome sequence contains 140 genes that has 94 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. There are 27 genes in each IR regions that includes 16 protein-coding genes species (rps19, rpl2, rpl23, ycf2, rps12, rps7, ndhB, ycf1, rps15, ndhH, ndhA, ndhI, ndhG, ndhE, psaC, and ndhD), 7 tRNA genes species (trnI-CAU, trnL-CAA, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, and trnN-GUU), and 4 rRNA genes species (rrn16, rrn23, rrn4.5, and rrn5).
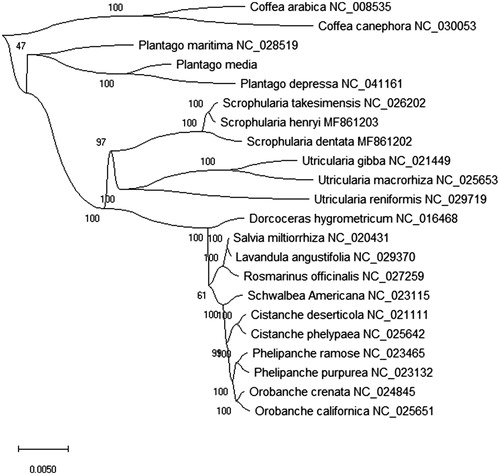
For the phylogenetic analysis, we study the phylogenetic relationship of P. media with other 21 plant species chloroplast genomes sequence. We constructed the phylogenetic tree by the maximum-likelihood (ML) method and analyzed the phylogenetic relationship using MEGA X (Kumar et al. Citation2018) with best model. The phylogenetic tree were inferred with strong support and used the bootstrap values from 2000 replicates at all the nodes. The phylogenetic tree was drawn and edited by MEGA X. In phylogenetic analysis, the result showed that P. media is closely related to P. depressa of the family Plantaginaceae in the phylogenetic relationship (). This research is great significance for the collection of traditional Chinese medicine resources in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Asaf S, Khan AL, Lubna A, Khan A, Khan G, Lee IJ, Al-Harrasi A. 2020. Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Scientific Reports. 10:3881.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.,
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715–2164.
- Ti S. 2015. Healing herbs: a beginner’s guide to identifying, foraging, and using medicinal plants/more than 100 remedies from 20 of the most healing plants. Beverly, MA: Fair Winds Press.