Abstract
Epimedium simplicifolium Ying is a rare and endangered species endemic to Guizhou, China. In this study, the complete chloroplast (cp) genome of E. simplicifolium was sequenced. The total cp genomes size was 158,745 bp in length, with 38.7% of GC content, including four distinct regions: the large single-copy region (LSC, 88,413 bp), small single-copy region (SSC, 15,386 bp), and a pair of inverted repeat regions (IRs, each for 27,473 bp). The whole cp genome of E. simplicifolium encoded 131 unique genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis with the previously reported cp genomes showed that all the samples of Epimedium are grouped into one group, which can be easily separated from the other genera of the Berberidaceae, and the E. simplicifolium is a separate species from the rest of the Epimedium. This plastid genome is the first report for the E. simplicifolium and will be useful data for developing markers for further studies on resolving the relationship within the Berberidaceae.
Berberidaceae (Ranales) is mainly distributed in the northern temperate and subtropical alpine regions, with about 650 species in 17 genera in worldwide, of which about 320 species in 11 genera are distributed in China and are famous for its medicinal value. Epimdium, Sinopodophyllum, Mahonia and Dysosma have the therapeutic effects of nourishing kidney, anticancer, strengthening bones and relieving rheumatism, and have a long history of medical use in traditional Chinese medicine (TCM) (Ying et al. Citation2011). In recent years, the research on Berberidaceae family has attracted more attention.
Epimdium simplicifolium, a rare and endangered species endemic to GuiZhou, China, which belongs to Epimdium (Berberidaceae), is a new species discovered by professor Ying Junsheng in 1970s, and only distributed in a small area in the Wuchuan and Suiyang county of Guizhou, China, with a very small population and urgently needs to be protected (He Citation2014), it is distinguished from other species by its simple and abaxially densely sericeous leaves. In this study, we sequenced the cp genomes of E. simplicifolium, aiming to provide valuable genetic information for the study of evolutionary dynamics and conservation.
In the resent study, the fresh young leaves of E. simplicifolium were collected from the Suiyang county, Guizhou Province, China, (N28°14′25.07″, E107°11′49.48″), and a voucher specimen (with collection numbers of YFL_20180343) has been deposited in the Herbarium of Guizhou University of Traditional Chinese Medicine (GZYGH), Guizhou, China. Total DNA was extracted from the fresh leaves of E. simplicifolium by the modified CTAB method (Doyle and Doyle Citation1987). Sequencing was carried out on the Illumina HiSeq X-Ten to generate approximately 3GB 150 bp reads at Beijing Genomics Institute (BGI, Wuhan, China). The filtered reads were assembled into a complete cp genome by the program GetOrganelle v 1.5 (Jin et al. Citation2018). In this pipeline, the complete cp genome reads were extracted from total genomic reads and were subsequently assembled using SPAdes version 3.10 (Bankevich et al. Citation2012). The genes were annotated using PGA (Qu et al. Citation2019) and Geneious 11.0.3 (Kearse et al. Citation2012) with the published complete cp genome of E. lishihchenii (GenBank accession number: NC_029944) as the reference. Transfer RNAs (tRNAs) were confirmed by their specific structure predicted by tRNAscan-SE 2.0 (Lowe and Chan Citation2016).
The complete cp genome of E. simplicifolium (GenBank accession number: MN939632) is 158,745 bp in length, with 38.7% of GC content, displaying a quadripartite structure that contains the large single-copy region (LSC, 88,413 bp), small single-copy region (SSC, 15,386 bp), and a pair of inverted repeat regions (IRs, each for 27,473 bp). There are 131 unique genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. A total of 20 genes (trnK-UUU, trnI-GAU × 2, trnA-UGC × 2, trnG-UCC, trnV-UAC, trnL-UAA, rpoC1, ndhB × 2, ndhA, rpl16, rpl2, petB, atpF, petD, rps16, rps12 × 2) contained 1 intron, and 2 genes (clpP, ycf3) contained 2 introns.
In order to explore the phylogenetic relationship of E. simplicifolium, we downloaded the whole cp genomes of 25 species from the NCBI GenBank database. Maximum likelihood (ML) analyses were performed using RAxML software (Stamatakis Citation2014).
The phylogenetic analysis was carried out using the complete cp genome sequences of E. simplicifolium, 16 reported Epimedium and 9 species from other genera of the Berberidaceae ().
Figure 1. Phylogenetic tree produced by maximum likelihood (ML) analysis based on chloroplast genome sequences from 26 species of Berberidaceae. Shown next to the nodes are bootstrap support values based on 1000 replicates.
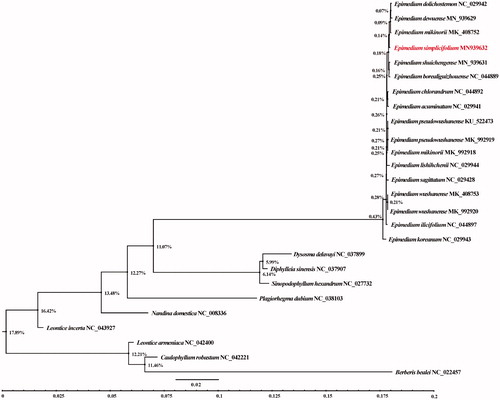
The results showed that all the 16 species of the Epimedium clustered into a supported branch, were separated from other species of Berberidaceae. Epimedium simplicifolium into a supported branch, is separated from other species of Epimedium. This cp genome is the first report for the E. simplicifolium and will be useful data for developing markers for further studies on resolving the relationship within the Berberidaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study has been deposited in GenBank with accession numbers MN939632.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- He SZ. 2014. Color map of the genus Epimedium in China. Guiyang: Guizhou Science and Technology Press.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular cp genome using genome skimming data. BioRxiv. :256479. doi:10.1101/256479
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15(50).
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Ying J, Boufford DE, Brach AR. 2011. Berberidaceae. Flora China. 19:714–800.
