Abstract
In this study, we report the complete mitochondrial genome of the iridescent blue-green damselfish, Chromis viridis (Perciformes, Pomacentridae), widely distributed species in the Indo-Pacific regions. The complete mitogenome of C. viridis was 16,894 bp long and was typical of Pomacentrinae mitogenomes in genomic content and structure, as the entire mitogenome contained 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 putative control region. Phylogenetic analysis of C. viridis within the family Pomacentridae based on the concatenated nucleotide sequences of entire PCGs confirmed the sister relationship among damselfish and anemonefish. This mitogenome sequence will provide a useful resource for understanding C. viridis-specific cryptic diversification and phylogeographic relationships in Pomacentridae.
The family Pomacentridae comprises numerous fish species with complex diversity and phylogenetic history. The iridescent blue-green damselfish inhabiting tropical and subtropical reefs or near-reef, Chromis viridis (Cuvier 1830), also known as blue puller belongs to the subfamily Pomacentrinae and is distributed across the Indo-West Pacific regions (Lieske and Myers Citation1994). The evolutionary process of blue-green damselfishes is complex due to the morphometric parameters and the overlap of the geographical distribution of the black-axil chromis Chromis atripectoralis. Previously, it was suggested that C. viridis and C. atripectoralis are sibling species based on phylogenetic results using mitochondrial ATPase8, ATPase6, and cytochrome b (Cytb) genes (Quenouille et al. Citation2004). Subsequently, extensive analysis using the mitochondrial control region of 88 blue-green damselfishes from Indonesia, the Philippines, Red Sea and the Great Barrier Reef confirmed C. viridis and C. atripectoralis as two species (Froukh and Kochzius Citation2008). Accumulation of cryptic lineages has been consistently suggested in C. viridis (Froukh and Kochzius Citation2008; Messmer et al. Citation2012). Recently, it was identified that two clades of C. viridis (C. viridis A and B) are cryptic taxa based on the substantial genetic divergence observed in both mtDNA (Cytb) and nuclear loci (Rag2) (Liu et al. Citation2019). Despite its wide distribution, the phylogenetic relationship and the taxonomic status of this species is still unclear due to the absence of whole mitochondrial genome information.
Here, we assembled the entire mitogenome of C. viridis (Accession no. MT199208) by employing Illumina MiSeq platform (Illumina, San Diego, CA, USA). A small tissue sample (0.22 g as wet weight) was isolated from the muscle of C. viridis that was originally collected at East China Sea (30°45′33.0″N, 126°22′10.2″E) in July 2013. The voucher specimen was deposited in the Research Institute of Basic Sciences of Incheon National University (Specimen ID: 2008-Pomacentridae043). Extraction of the total genomic DNA was performed using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA), followed by sequencing library generation with TruSeq RNA Sample Preparation Kit according to the manufacturer’s instructions (Illumina) and paired-end sequencing was conducted on Illumina MiSeq platform (Illumina) at Phyzen (Seoul, South Korea). CLC Assembly Cell package (version 4.2.1) with the CLC de novo assemble algorithm was used for assembly. Additional PCR procedure and Sanger sequencing were conducted to confirm the nucleotide sequence of putative control region. Entire C. viridis mitogenome was annotated using the MITOS web-based software (Bernt et al. Citation2013) and detailed annotation were conducted with NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
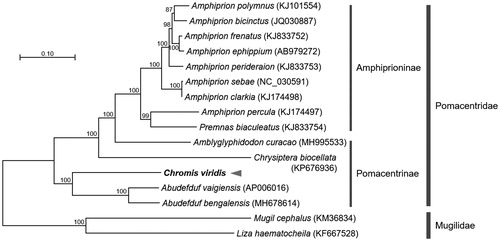
The complete mitochondrial genome of C. viridis was 16,894 bp in length and contained a typical set of 13 PCGs, 22 tRNAs, 2 rRNAs, and 1 control region located in the arrangement typical of Pomacentrinae mitogenomes. A phylogenetic analysis was constructed using the concatenated set of entire 13 PCGs of C. viridis mitogenome including 15 published mitogenomes from Pomacentridae and Mugilidae (ourgroup) (). We used JModelTest ver. 2.1.10 (Darriba et al. Citation2012) to select the best substitution model and a (HKY + G + I) substitution model was employed to construct a maximum-likelihood (ML) method in the PhyML 2.4.5 (Guindon and Gascuel Citation2003) with 1000 bootstrap replicates. Phylogenetic relationship showed that C. viridis grouped together with other representatives of subfamily Pomacentrinae, and a highly supported clade has recovered between genus Amphiprion, Amblyglyphidodon, Chrysiptera, and Abudefduf (Floeter et al. Citation2018). In conclusion, the complete C. viridis mitogenome will provide useful information to elucidate cryptic characteristics, phylogeography, and ecological niche of the family Pomacentridae.
Figure 1. Maximum-likelihood (ML) phylogeny of 15 species of the family Pomacentridae based on the concatenated nucleotide sequences of entire protein-coding genes (PCGs). Two species from the family Mugilidae were used as outgroup. Numbers on the branches indicate ML bootstrap percentages (1000 replicates).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772
- Floeter SR, Bender MG, Siqueira AC, Cowman PF. 2018. Phylogenetic perspectives on reef fish functional traits. Biol Rev. 93(1):131–151.
- Froukh T, Kochzius M. 2008. Species boundaries and evolutionary lineages in the blue green damselfishes Chromis viridis and Chromis atripectoralis (Pomacentridae). J Fish Biol. 72(2):451–457.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systemat Biol. 52(5):696–704.
- Lieske E, Myers R. 1994. Collins pocket guide. Coral reef fishes. Indo-Pacific & Caribbean including the Red Sea. New York: Haper Collins Publishers.
- Liu SYV, Tuanmu MN, Rachmawati R, Mahardika GN, Barber PH. 2019. Integrating phylogeographic and ecological niche approaches to delimitating cryptic lineages in the blue-green damselfish (Chromis viridis). PeerJ. 7:e7384.
- Messmer V, Jones GP, Munday PL, Planes S. 2012. Concordance between genetic and species diversity in coral reef fishes across the Pacific Ocean biodiversity gradient. Evolution. 66(12):3902–3917.
- Quenouille B, Bermingham E, Planes S. 2004. Molecular systematics of the damselfishes (Teleostei: Pomacentridae): Bayesian phylogenetic analyses of mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 31(1):66–88.
