Abstract
In this study, we report on the complete mitochondrial genome of the fire goby, Nemateleotris magnifica (Perciformes, Gobiidae), a small attractive ornamental fish distributed in the Indo-Pacific region. The complete mitogenome of N. magnifica was 16,502 bp long and was typical of genus Nemateleotris mitogenomes in genomic content and structure, as the entire mitogenome contained 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 control region. The phylogenetic analysis within gobiine-like and gobionelline-like gobiids based on the concatenated nucleotide sequences of the entire 13 PCGs confirmed the involvement in the gobiine-like group and the sister relationship with Nemateleotris decora. This mitogenome sequence will provide a useful resource for comparative analysis of the phylogenetic distance in gobiine-like gobiids.
The Gobiidae family is one of the largest vertebrate families with a world-wide distribution (Thacker Citation2009). They comprise predominantly bottom-dwelling species and have both tropical and temperate habitats with a variety in morphology, ecology, and behavior (Agorreta et al. Citation2013). They are generally benthic and are distributed in freshwater, estuary, and ocean, in particular coral reefs (Thacker and Roje Citation2011). Gobiidae comprise two representative clades, the gobiine-like and the gobionelline-like gobiids based on phylogenetic analysis using nuclear and mitochondrial sequences (Agorreta et al. Citation2013; Tornabene et al. Citation2013; Adrian-Kalchhauser et al. Citation2017). The fire goby, Nemateleotris magnifica is a popular ornamental species which is recognized by the distinct elongated first dorsal fin, bright yellow snout, and unique body color shaded from white/silver to orange-red posteriorly. It has a vast distribution in tropical marine waters of the Indo-Pacific and Central Pacific. Since several interesting issues such as diversification, morphometry, phylogeography, and evolutionary history have been highlighted in Gobiidae (Agorreta et al. Citation2013), the accumulation of gobiid mitogenome information will be useful to understand their molecular phylogenetic distance and complex diversity.
In this study, we assembled the entire mitogenome of N. magnifica (Accession no. MT199210) by employing Illumina MiSeq platform (Illumina, San Diego, CA, USA). Muscle tissue (0.28 g as wet weight) was isolated from an individual of N. magnifica that was originally collected at East China Sea (30°45′33.0″N 126°22′10.2″E) in July 2013. The voucher specimen was registered with a specimen ID: 2013-gobiidae012 and deposited in the Research Institute of Basic Sciences of Incheon National University (Incheon, South Korea). Isolation of the total genomic DNA was conducted using DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany), followed by sequencing library generation with TruSeq RNA Sample Preparation Kit based on the manufacturer’s instructions (Illumina). Paired-end sequencing was conducted on Illumina MiSeq platform (Illumina) at Phyzen (Seoul, South Korea). CLC Assembly Cell package (version 4.2.1) with the CLC de novo assemble algorithm was employed for assembly. Additional PCR procedures and Sanger sequencing was conducted to confirm the nucleotide sequence of the control region. The whole N. magnifica mitogenome was annotated by using the MITOS web-based software (Bernt et al. Citation2013) and the results were confirmed with NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
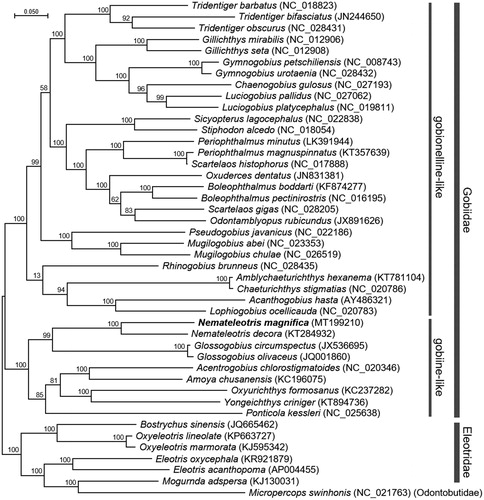
The complete mitochondrial genome of N. magnifica was 16,502 bp in length and contained a typical set of 13 PCGs, 22 tRNAs, 2 rRNAs, and 1 control region. A phylogenetic analysis was constructed using the concatenated set of entire 13 PCGs of N. magnifica mitogenome including 28 published mitogenomes from gobionelline-like and 8 published mitogenomes from gobiine-like gobiids (). Overall, whole mitochondrial genome phylogenies were similar to that of Adrian-Kalchhauser et al (Citation2017), although slight differences were observed, presumably due to the incorporation of 12 PCGs for their analysis. In addition, species having incomplete sequences or unverified mitogenomes were excluded from phylogeny analysis in this study. The phylogenetic relationship showed that N. magnifica formed a cluster with Nemateleotris decora and involved together in gobiine-like gobiids. Since relatively small numbers of whole mitogenomes in gobiine-like gobiids have been registered in NCBI compared to those of gobionelline-like gobiids, this information will be an essential resource to elucidate the phylogenetic relationship, geographical distribution, and evolution of gobiine-like gobiids.
Figure 1. Maximum-likelihood (ML) phylogeny of 28 species of gobionelline-like and 8 species of gobiine-like gobiids based on the 13 concatenated nucleotide sequences of the entire protein-coding genes (PCGs). Six species from the family Eleotridae and a species from the family Odontobutidae were incorporated to build a reliable phylogeny tree. Numbers on the branches indicate ML bootstrap percentages (1000 replicates). DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Adrian-Kalchhauser I, Svensson O, Kutschera VE, Alm Rosenblad M, Pippel M, Winkler S, Schloissnig S, Blomberg A, Burkhardt-Holm P. 2017. The mitochondrial genome sequences of the round goby and the sand goby reveal patterns of recent evolution in gobiid fish. BMC Genomics. 18(1):177.
- Agorreta A, San Mauro D, Schliewen U, Van Tassell JL, Kovačić M, Zardoya R, Rüber L. 2013. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol Phylogenet Evol. 69(3):619–633.
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Thacker CE, Roje DM. 2011. Phylogeny of Gobiidae and identification of gobiid lineages. Syst Biodiv. 9(4):329–347.
- Thacker CE. 2009. Phylogeny of Gobioidei and placement within acanthomorpha, with a new classification and investigation of diversification and character evolution. Copeia. 2009(1):93–104.
- Tornabene L, Chen Y, Pezold F. 2013. Gobies are deeply divided: phylogenetic evidence from nuclear DNA (Teleostei: Gobioidei: Gobiidae). Syst Biodivers. 11(3):345–361.
